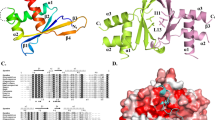
Abstract
The male-specific lethal (MSL) complex is required for dosage compensation in Drosophila melanogaster, and analogous complexes exist in mammals. We report structures of binary complexes of mammalian MSL3 and the histone acetyltransferase (HAT) MOF with consecutive segments of MSL1. MSL1 interacts with MSL3 as an extended chain forming an extensive hydrophobic interface, whereas the MSL1-MOF interface involves electrostatic interactions between the HAT domain and a long helix of MSL1. This structure provides insights into the catalytic mechanism of MOF and enables us to show analogous interactions of MOF with NSL1. In Drosophila, selective disruption of Msl1 interactions with Msl3 or Mof severely affects Msl1 targeting to the body of dosage-compensated genes and several high-affinity sites, without affecting promoter binding. We propose that Msl1 acts as a scaffold for MSL complex assembly to achieve specific targeting to the X chromosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Straub, T. & Becker, P.B. Dosage compensation: the beginning and end of generalization. Nat. Rev. Genet. 8, 47–57 (2007).
Heard, E. & Disteche, C.M. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 20, 1848–1867 (2006).
Mendjan, S. & Akhtar, A. The right dose for every sex. Chromosoma 116, 95–106 (2007).
Gelbart, M.E. & Kuroda, M.I. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136, 1399–1410 (2009).
Smith, E.R. et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25, 9175–9188 (2005).
Marín, I. Evolution of chromatin-remodeling complexes: comparative genomics reveals the ancient origin of “novel” compensasome genes. J. Mol. Evol. 56, 527–539 (2003).
Mendjan, S. et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21, 811–823 (2006).
Taipale, M. et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25, 6798–6810 (2005).
Alekseyenko, A.A. et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609 (2008).
Smith, E.R. et al. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20, 312–318 (2000).
Akhtar, A. & Becker, P.B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000).
Hilfiker, A., Hilfiker-Kleiner, D., Pannuti, A. & Lucchesi, J.C. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16, 2054–2060 (1997).
Scott, M.J., Pan, L.L., Cleland, S.B., Knox, A.L. & Heinrich, J. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J. 19, 144–155 (2000).
Morales, V. et al. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 23, 2258–2268 (2004).
Morales, V., Regnard, C., Izzo, A., Vetter, I. & Becker, P.B. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol. Cell. Biol. 25, 5947–5954 (2005).
Sural, T.H. et al. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat. Struct. Mol. Biol. 15, 1318–1325 (2008).
Akhtar, A., Zink, D. & Becker, P.B. Chromodomains are protein-RNA interaction modules. Nature 407, 405–409 (2000).
Li, X., Wu, L., Corsa, C.A., Kunkel, S. & Dou, Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 36, 290–301 (2009).
Raja, S.J. et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell 38, 827–841 (2010).
Yan, Y., Barlev, N.A., Haley, R.H., Berger, S.L. & Marmorstein, R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6, 1195–1205 (2000).
Rojas, J.R. et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 (1999).
Wang, L., Tang, Y., Cole, P.A. & Marmorstein, R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747 (2008).
Yan, Y., Harper, S., Speicher, D.W. & Marmorstein, R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9, 862–869 (2002).
Berndsen, C.E., Albaugh, B.N., Tan, S. & Denu, J.M. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46, 623–629 (2007).
Burke, T.W., Cook, J.G., Asano, M. & Nevins, J.R. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276, 15397–15408 (2001).
Zhang, P. et al. The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Sci. 15, 2423–2434 (2006).
Bowman, B.R. et al. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure 14, 151–158 (2006).
Buscaino, A. et al. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol. Cell 11, 1265–1277 (2003).
Kelley, R.L. et al. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98, 513–522 (1999).
Sanchez-Weatherby, J. et al. Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallogr. D Biol. Crystallogr. 65, 1237–1246 (2009).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Davis, I.W., Murray, L.W., Richardson, J.S. & Richardson, D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
Brunger, A.T., DeLaBarre, B., Davies, J.M. & Weis, W.I. X-ray structure determination at low resolution. Acta Crystallogr. D Biol. Crystallogr. 65, 128–133 (2009).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank K. Dzeyk of EMBL Heidelberg and I. Berard of the Institut de Biologie Structurale (IBS) Grenoble for their help with mass spectrometry; the High Throughput Crystallisation Laboratory (HTX) group of EMBL Grenoble for performing initial screening crystallization trials; M. Jamin's group for help with the MALLS experiment; the European Synchrotron Radiation Facility (ESRF)-EMBL Joint Structural Biology Group for access to and assistance on the ESRF synchrotron beamlines; P. Tropberger from R. Schneider's laboratory (Max Planck Institute of Immunology, Freiburg) for providing nucleosomes; and D. Panne and members of both laboratories for critical reading of the manuscript. E.H. is a Darwin trust fellow.
Author information
Authors and Affiliations
Contributions
J.K., E.H., S.C. and A.A. designed the project and wrote the manuscript; J.K. and M.L. produced and crystallized the proteins; J.K. solved, refined and analyzed the structures, prepared and tested the mutants in vitro, and carried out HAT assays on the H4 peptide; J.S.-W. and J.K. performed dehydration of MSL1–MSL3 crystals; H.H. performed HAT assays on nucleosomes; and E.H. carried out all in vivo experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Table 1 and Supplementary Methods (PDF 1551 kb)
Rights and permissions
About this article
Cite this article
Kadlec, J., Hallacli, E., Lipp, M. et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol 18, 142–149 (2011). https://doi.org/10.1038/nsmb.1960
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1960
This article is cited by
-
Structural and functional insights into the epigenetic regulator MRG15
Acta Pharmacologica Sinica (2024)
-
Two distinct males absent on the first (MOF)-containing histone acetyltransferases are involved in the epithelial–mesenchymal transition in different ways in human cells
Cellular and Molecular Life Sciences (2022)
-
Interaction of Male Specific Lethal complex and genomic imbalance on global gene expression in Drosophila
Scientific Reports (2021)
-
Disruption of the MSL complex inhibits tumour maintenance by exacerbating chromosomal instability
Nature Cell Biology (2021)
-
Acquisition of a side population fraction augments malignant phenotype in ovarian cancer
Scientific Reports (2019)