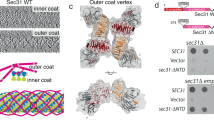
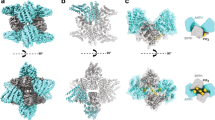
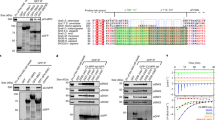
Abstract
Multisubunit tethering complexes participate in the process of vesicle tethering—the initial interaction between transport vesicles and their acceptor compartments. TRAPPII (named for transport protein particle II) is a highly conserved tethering complex that functions in the late Golgi apparatus and consists of all of the subunits of TRAPPI and three additional, specific subunits. We have purified native yeast TRAPPII and characterized its structure and subunit organization by single-particle EM. Our data show that the nine TRAPPII components form a core complex that dimerizes into a three-layered, diamond-shaped structure. The TRAPPI subunits assemble into TRAPPI complexes that form the outer layers. The three TRAPPII-specific subunits cap the ends of TRAPPI and form the middle layer, which is responsible for dimerization. TRAPPII binds the Ypt1 GTPase and probably uses the TRAPPI catalytic core to promote guanine nucleotide exchange. We discuss the implications of the structure of TRAPPII for coat interaction and TRAPPII-associated human pathologies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Whyte, J.R. & Munro, S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115, 2627–2637 (2002).
Sztul, E. & Lupashin, V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett. 583, 3770–3783 (2009).
Cai, H., Reinisch, K. & Ferro-Novick, S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671–682 (2007).
Kümmel, D. & Heinemann, U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr. Protein Pept. Sci. 9, 197–209 (2008).
Sacher, M., Kim, Y.G., Lavie, A., Oh, B.H. & Segev, N. The TRAPP complex: insights into its architecture and function. Traffic 9, 2032–2042 (2008).
Lynch-Day, M.A. et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107, 7811–7816 (2010).
Sacher, M. et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell 7, 433–442 (2001).
Jones, S., Newman, C., Liu, F. & Segev, N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell 11, 4403–4411 (2000).
Wang, W., Sacher, M. & Ferro-Novick, S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151, 289–296 (2000).
Cai, H. et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature 445, 941–944 (2007).
Kim, Y.G. et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 127, 817–830 (2006).
Cai, Y. et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133, 1202–1213 (2008).
Montpetit, B. & Conibear, E. Identification of the novel TRAPP associated protein Tca17. Traffic 10, 713–723 (2009).
Scrivens, P.J. et al. TRAPPC2L is a novel, highly conserved TRAPP-interacting protein. Traffic 10, 724–736 (2009).
Cai, H., Zhang, Y., Pypaert, M., Walker, L. & Ferro-Novick, S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J. Cell Biol. 171, 823–833 (2005).
Yamasaki, A. et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell 20, 4205–4215 (2009).
Morozova, N. et al. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8, 1263–1269 (2006).
Gedeon, A.K. et al. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat. Genet. 22, 400–404 (1999).
Gedeon, A.K. et al. The molecular basis of X-linked spondyloepiphyseal dysplasia tarda. Am. J. Hum. Genet. 68, 1386–1397 (2001).
Mir, A. et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am. J. Hum. Genet. 85, 909–915 (2009).
Mochida, G.H. et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am. J. Hum. Genet. 85, 897–902 (2009).
Philippe, O. et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am. J. Hum. Genet. 85, 903–908 (2009).
Kampmann, M. & Blobel, G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 16, 782–788 (2009).
Cheng, Y. et al. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 355, 1048–1065 (2006).
Radermacher, M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron Microsc. Tech. 9, 359–394 (1988).
Sacher, M., Barrowman, J., Schieltz, D., Yates, J.R. III & Ferro-Novick, S. Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 79, 71–80 (2000).
Ren, Y. et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell 139, 1119–1129 (2009).
Lees, J.A., Yip, C.K., Walz, T. & Hughson, F.M. Molecular organization of the COG vesicle tethering complex. Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1917 (24 October 2010).
Cox, R., Chen, S.H., Yoo, E. & Segev, N. Conservation of the TRAPPII-specific subunits of a Ypt/Rab exchanger complex. BMC Evol. Biol. 7, 12 (2007).
Kümmel, D., Oeckinghaus, A., Wang, C., Krappmann, D. & Heinemann, U. Distinct isocomplexes of the TRAPP trafficking factor coexist inside human cells. FEBS Lett. 582, 3729–3733 (2008).
Jedd, G., Richardson, C., Litt, R. & Segev, N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J. Cell Biol. 131, 583–590 (1995).
Sclafani, A. et al. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic 11, 520–532 (2010).
Jang, S.B. et al. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J. Biol. Chem. 277, 49863–49869 (2002).
Choi, M.Y. et al. Biochemical consequences of sedlin mutations that cause spondyloepiphyseal dysplasia tarda. Biochem. J. 423, 233–242 (2009).
Longtine, M.S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 (1999).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Li, Z., Hite, R.K., Cheng, Y. & Walz, T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J. Electron Microsc. (Tokyo) 59, 53–63 (2010).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Böttcher, B., Wynne, S.A. & Crowther, R.A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91 (1997).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Christoforidis, S. & Zerial, M. Purification and identification of novel Rab effectors using affinity chromatography. Methods 20, 403–410 (2000).
Murata, T. et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 (2006).
Acknowledgements
We thank J. Nicols for technical assistance with purification and data processing and Z. Li for assistance with microscopy and image processing. C.K.Y. acknowledges fellowships from the Jane Coffin Childs Memorial Fund and the Canadian Institutes of Health Research. T.W. is an investigator in the Howard Hughes Medical Institute. The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from US National Institutes of Health (NIH) grant PO1 GM62580 (to S.C. Harrison). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Author information
Authors and Affiliations
Contributions
C.K.Y. and T.W. conceived the experiments. C.K.Y. performed all experiments. J.B. assisted in generating GFP-labeled yeast strains and data processing. C.K.Y. and T.W. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 2477 kb)
Rights and permissions
About this article
Cite this article
Yip, C., Berscheminski, J. & Walz, T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol 17, 1298–1304 (2010). https://doi.org/10.1038/nsmb.1914
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1914
This article is cited by
-
The substrate specificity of the human TRAPPII complex’s Rab-guanine nucleotide exchange factor activity
Communications Biology (2020)
-
Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles
Scientific Reports (2017)
-
Chaperoning SNARE assembly and disassembly
Nature Reviews Molecular Cell Biology (2016)
-
Identification of conserved, centrosome-targeting ASH domains in TRAPPII complex subunits and TRAPPC8
Cilia (2014)
-
Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation
Cellular and Molecular Life Sciences (2014)