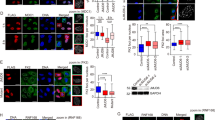
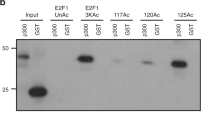
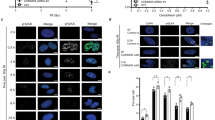
Abstract
DNA double-strand break (DSB) repair occurs within chromatin and can be modulated by chromatin-modifying enzymes. Here we identify the related human histone deacetylases HDAC1 and HDAC2 as two participants in the DNA-damage response. We show that acetylation of histone H3 Lys56 (H3K56) was regulated by HDAC1 and HDAC2 and that HDAC1 and HDAC2 were rapidly recruited to DNA-damage sites to promote hypoacetylation of H3K56. Furthermore, HDAC1- and 2-depleted cells were hypersensitive to DNA-damaging agents and showed sustained DNA-damage signaling, phenotypes that reflect defective DSB repair, particularly by nonhomologous end-joining (NHEJ). Collectively, these results show that HDAC1 and HDAC2 function in the DNA-damage response by promoting DSB repair and thus provide important insights into the radio-sensitizing effects of HDAC inhibitors that are being developed as cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Harper, J.W. & Elledge, S.J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
Huertas, P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 17, 11–16 (2010).
Gottlieb, T.M. & Jackson, S.P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72, 131–142 (1993).
Mahaney, B.L., Meek, K. & Lees-Miller, S.P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417, 639–650 (2009).
Downs, J.A., Nussenzweig, M.C. & Nussenzweig, A. Chromatin dynamics and the preservation of genetic information. Nature 447, 951–958 (2007).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Misteli, T. & Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10, 243–254 (2009).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Bonner, W.M. et al. γH2AX and cancer. Nat. Rev. Cancer 8, 957–967 (2008).
Stucki, M. & Jackson, S.P. γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.) 5, 534–543 (2006).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Huen, M.S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Kolas, N.K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Stewart, G.S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Escargueil, A.E., Soares, D.G., Salvador, M., Larsen, A.K. & Henriques, J.A. What histone code for DNA repair? Mutat. Res. 658, 259–270 (2008).
Corpet, A. & Almouzni, G. A histone code for the DNA damage response in mammalian cells? EMBO J. 28, 1828–1830 (2009).
Murr, R. et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91–99 (2006).
Tjeertes, J.V., Miller, K.M. & Jackson, S.P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009).
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009).
Das, C., Lucia, M.S., Hansen, K.C. & Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Yang, B., Zwaans, B.M., Eckersdorff, M. & Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 (2009).
Glozak, M.A. & Seto, E. Histone deacetylases and cancer. Oncogene 26, 5420–5432 (2007).
Yang, X.J. & Seto, E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 (2008).
Haigis, M.C. & Sinclair, D.A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 (2010).
Smith, K.T. & Workman, J.L. Histone deacetylase inhibitors: anticancer compounds. Int. J. Biochem. Cell Biol. 41, 21–25 (2009).
Camphausen, K. & Tofilon, P.J. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J. Clin. Oncol. 25, 4051–4056 (2007).
Iacovoni, J.S. et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 (2010).
d'Adda di Fagagna, F. et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 (2003).
d'Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008).
Bartkova, J. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006).
Serrano, M., Lin, A.W., McCurrach, M.E., Beach, D. & Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Xie, W. et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell 33, 417–427 (2009).
Willis-Martinez, D., Richards, H.W., Timchenko, N.A. & Medrano, E.E. Role of HDAC1 in senescence, aging, and cancer. Exp. Gerontol. 45, 279–285 (2010).
Bandyopadhyay, D. et al. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 62, 6231–6239 (2002).
Yuan, J., Pu, M., Zhang, Z. & Lou, Z. Histone H3–K56 acetylation is important for genomic stability in mammals. Cell Cycle 8, 1747–1753 (2009).
Miller, K.M., Maas, N.L. & Toczyski, D.P. Taking it off: regulation of H3 K56 acetylation by Hst3 and Hst4. Cell Cycle 5, 2561–2565 (2006).
Luo, J. et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137–148 (2001).
Cheng, H.L. et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100, 10794–10799 (2003).
Dovey, O.M., Foster, C.T. & Cowley, S.M. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA 107, 8242–8247 (2010).
Pommier, Y. et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532, 173–203 (2003).
Uematsu, N. et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 177, 219–229 (2007).
Mari, P.O. et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 103, 18597–18602 (2006).
Higashiura, M., Shimizu, Y., Tanimoto, M., Morita, T. & Yagura, T. Immunolocalization of Ku-proteins (p80/p70): localization of p70 to nucleoli and periphery of both interphase nuclei and metaphase chromosomes. Exp. Cell Res. 201, 444–451 (1992).
Bird, A.W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415 (2002).
Jazayeri, A., McAinsh, A.D. & Jackson, S.P. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101, 1644–1649 (2004).
Xhemalce, B. et al. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe . J. Biol. Chem. 282, 15040–15047 (2007).
Li, B., Carey, M. & Workman, J.L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009).
Galanty, Y. et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 (2009).
Roberts, S.A. & Ramsden, D.A. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J. Biol. Chem. 282, 10605–10613 (2007).
Kim, G.D. et al. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J. Biol. Chem. 274, 31127–31130 (1999).
Rimkus, S.A. et al. Mutations in String/CDC25 inhibit cell cycle re-entry and neurodegeneration in a Drosophila model of Ataxia telangiectasia. Genes Dev. 22, 1205–1220 (2008).
Goodarzi, A.A. et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 (2008).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Gorgoulis, V.G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Fraga, M.F. et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 (2005).
Ocker, M. & Schneider-Stock, R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 39, 1367–1374 (2007).
Acknowledgements
We thank all members of the Jackson laboratory for help and support, P. Huertas, A. Kaidi and B. Xhemalce for reading of the manuscript, B. Xhemalce for assistance with ChIP, F. d'Adda di Fagagna for OIS cells, A. Meier for replicative senescent BJ cells, P. Frit for GFP-KU70 and NEB Biolabs for AsiSI genomic DNA. K.M.M. is funded by a Wellcome Trust project grant (086861/Z/08/Z). J.V.T. is supported by a Biotechnology and Biological Sciences Research Council Cooperative Award in Science and Engineering studentship with KuDOS Pharmaceuticals. G.L.'s research is funded by grants from Association pour la Recherche sur le Cancer, the Centre National de la Recherche Scientifique and PRES-University of Toulouse. S.E.P. is funded by the Human Frontier Science Program Organization. S.B. is funded by Cancer Research UK and an EMBO Long-Term Fellowship. Research in the S.P.J. laboratory is also supported by the European Community (EU Projects DNA Repair (LSHG-CT-2005-512113) and GENICA) and by core infrastructure provided by Cancer Research UK and the Wellcome Trust. We thank F. d'Adda di Fagagna (IFOM-IEO) for providing OIS cells, A. Meier for replicative senescent BJ cells and P. Frit (CNRS) for GFP-Ku70.
Author information
Authors and Affiliations
Contributions
K.M.M., J.V.T., and J.C. performed experiments; G.L. provided the AsiSI-expressing cell line; S.E.P. provided initial observation of HDAC1 localization; S.B. created and assisted with cell lines expressing NHEJ-tagged factors; K.M.M., J.V.T. and S.P.J. designed and analyzed the experiments; K.M.M. and S.P.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Table 1 and Supplementary Figures 1–8 (PDF 1180 kb)
Rights and permissions
About this article
Cite this article
Miller, K., Tjeertes, J., Coates, J. et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17, 1144–1151 (2010). https://doi.org/10.1038/nsmb.1899
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1899
This article is cited by
-
CYLD induces high oxidative stress and DNA damage through class I HDACs to promote radiosensitivity in nasopharyngeal carcinoma
Cell Death & Disease (2024)
-
Prediction of hub genes and key pathways associated with the radiation response of human hematopoietic stem/progenitor cells using integrated bioinformatics methods
Scientific Reports (2023)
-
Recent advancement of HDAC inhibitors against breast cancer
Medical Oncology (2023)
-
The consequences of viral infection on host DNA damage response: a focus on SARS-CoVs
Journal of Genetic Engineering and Biotechnology (2022)
-
Lymphoid-specific helicase in epigenetics, DNA repair and cancer
British Journal of Cancer (2022)