Abstract
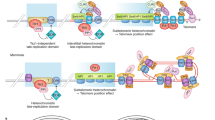
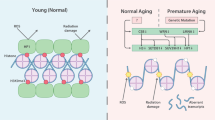
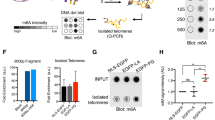
During replicative aging of primary cells morphological transformations occur, the expression pattern is altered and chromatin changes globally. Here we show that chronic damage signals, probably caused by telomere processing, affect expression of histones and lead to their depletion. We investigated the abundance and cell cycle expression of histones and histone chaperones and found defects in histone biosynthesis during replicative aging. Simultaneously, epigenetic marks were redistributed across the phases of the cell cycle and the DNA damage response (DDR) machinery was activated. The age-dependent reprogramming affected telomeric chromatin itself, which was progressively destabilized, leading to a boost of the telomere-associated DDR with each successive cell cycle. We propose a mechanism in which changes in the structural and epigenetic integrity of telomeres affect core histones and their chaperones, enforcing a self-perpetuating pathway of global epigenetic changes that ultimately leads to senescence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kaygun, H. & Marzluff, W.F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 25, 6879–6888 (2005).
Su, C. et al. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 23, 1133–1143 (2004).
Groth, A. et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 (2005).
Hoek, M. & Stillman, B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100, 12183–12188 (2003).
Das, C., Lucia, M.S., Hansen, K.C. & Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Tjeertes, J.V., Miller, K.M. & Jackson, S.P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009).
Bunz, F. et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998).
Wang, Z.F., Whitfield, M.L., Ingledue, T.C. III, Dominski, Z. & Marzluff, W.F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10, 3028–3040 (1996).
Whitfield, M.L. et al. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20, 4188–4198 (2000).
Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 (2000).
Gérard, A. et al. The replication kinase Cdc7-Dbf4 promotes the interaction of the p150 subunit of chromatin assembly factor 1 with proliferating cell nuclear antigen. EMBO Rep. 7, 817–823 (2006).
Juan, G., Hernando, E. & Cordon-Cardo, C. Separation of live cells in different phases of the cell cycle for gene expression analysis. Cytometry 49, 170–175 (2002).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Bernstein, B.E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005).
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003).
Francis, N.J., Follmer, N.E., Simon, M.D., Aghia, G. & Butler, J.D. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell 137, 110–122 (2009).
Goldstein, S. Replicative senescence: the human fibroblast comes of age. Science 249, 1129–1133 (1990).
Chen, E.S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008).
Kloc, A., Zaratiegui, M., Nora, E. & Martienssen, R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 18, 490–495 (2008).
Peters, A.H. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003).
Imai, S., Armstrong, C.M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008).
Chen, C.C. et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 (2008).
Hayashi, R. et al. Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol. 26, 91–99 (2007).
Jasencakova, Z. et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 (2010).
Harper, J.W. & Elledge, S.J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
Petrini, J.H. & Stracker, T.H. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13, 458–462 (2003).
Stracker, T.H., Couto, S.S., Cordon-Cardo, C., Matos, T. & Petrini, J.H. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol. Cell 31, 21–32 (2008).
Scaffidi, P. & Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 (2006).
Stewart, S.A. et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 99, 12606–12611 (2002).
Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309 (2007).
Grunstein, M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell 93, 325–328 (1998).
Bianchi, A. & Shore, D. Early replication of short telomeres in budding yeast. Cell 128, 1051–1062 (2007).
Meeks-Wagner, D. & Hartwell, L.H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43–52 (1986).
Steger, D.J. & Workman, J.L. Transcriptional analysis of purified histone acetyltransferase complexes. Methods 19, 410–416 (1999).
Harris, M.E. et al. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11, 2416–2424 (1991).
Dominski, Z., Zheng, L.X., Sanchez, R. & Marzluff, W.F. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19, 3561–3570 (1999).
Groth, A. et al. Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007).
Dang, W. et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 (2009).
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009).
Recht, J. et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 103, 6988–6993 (2006).
Li, Q. et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 (2008).
Maas, N.L., Miller, K.M., DeFazio, L.G. & Toczyski, D.P. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23, 109–119 (2006).
Schotta, G. et al. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002).
van Leeuwen, F., Gafken, P.R. & Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002).
Nishioka, K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 (2002).
Altaf, M. et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28, 1002–1014 (2007).
d'Adda di Fagagna, F. et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 (2003).
Denchi, E.L. & de Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 (2007).
Takai, H., Smogorzewska, A. & de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 (2003).
Dimitrova, N., Chen, Y.C., Spector, D.L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008).
Crabbe, L., Verdun, R.E., Haggblom, C.I. & Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306, 1951–1953 (2004).
Fodor, B.D. et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 (2006).
Verdun, R.E. & Karlseder, J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127, 709–720 (2006).
Azuara, V. Profiling of DNA replication timing in unsynchronized cell populations. Nat. Protoc. 1, 2171–2177 (2006).
Acknowledgements
We thank G. Almouzni (Institut Curie) for sharing antibodies to Asf1a and Asf1b and for helpful discussions. We are grateful to T. Jenuwein (Max Planck Institute of Immunobiology) for the gift of the H3K9, H3K27 and H4K20 series of antibodies, T. Hunter (Salk Institute) for cyclin antibodies, T. de Lange (The Rockefeller University) for advice on chronic damage protocols, J. Jaffe (Broad Institute) and S. Carr (Broad Institute) for access to the Broad Proteomics Platform, expert advice and helpful discussion. We also thank D. Chambers (Salk Institute) and J. Barrie (Salk Institute) for technical assistance with flow cytometry. We are grateful to members of the Karlseder lab for critical discussion of the manuscript. R.J.O'S. is supported by the George E. Hewitt Foundation for Medical Research, S.K. is supported by a postdoctoral fellowship of the Ernst Schering Research Foundation and the European Union, and J.K. acknowledges support by the US National Institutes of Health (RO1 GM06525 and RO1 AG025837).
Author information
Authors and Affiliations
Contributions
R.J.O'S. designed and carried out experiments and wrote the manuscript, S.K. did the MS analysis, S.L.S. provided advice and access to the MS facilities, and J.K. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 3315 kb)
Rights and permissions
About this article
Cite this article
O'Sullivan, R., Kubicek, S., Schreiber, S. et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17, 1218–1225 (2010). https://doi.org/10.1038/nsmb.1897
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1897
This article is cited by
-
Genetik, Epigenetik und Umweltfaktoren der Lebenserwartung – Welche Rolle spielt Nature-versus-Nurture beim Altern?
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2024)
-
Telomeres and aging: on and off the planet!
Biogerontology (2024)
-
Chromatin balances cell redox and energy homeostasis
Epigenetics & Chromatin (2023)
-
Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis
Nature (2023)
-
Differential protein expression in the hippocampi of resilient individuals identified by digital spatial profiling
Acta Neuropathologica Communications (2022)