Abstract
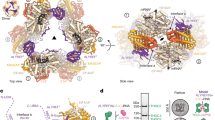
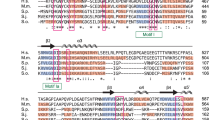
The binding of capped RNAs to the cap-binding complex (CBC) in the nucleus, and their dissociation from the CBC in the cytosol, represent essential steps in RNA processing. Here we show how the nucleocytoplasmic transport proteins importin-α and importin-β have key roles in regulating these events. As a first step toward understanding the molecular basis for this regulation, we determined a 2.2-Å resolution X-ray structure for a CBC–importin-α complex that provides a detailed picture for how importin-α binds to the CBP80 subunit of the CBC. Through a combination of biochemical studies, X-ray crystallographic information and small-angle scattering experiments, we then determined how importin-β binds to the CBC through its CBP20 subunit. Together, these studies enable us to propose a model describing how importin-β stimulates the dissociation of capped RNA from the CBC in the cytosol following its nuclear export.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
von der Haar, T., Gross, J.D., Wagner, G. & McCarthy, J.E.G. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11, 503–511 (2004).
Lewis, J.D. & Izaurralde, E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247, 461–469 (1997).
Cheng, H. et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127, 1389–1400 (2006).
Ishigaki, Y., Li, X., Serin, G. & Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001).
Lejeune, F., Ishigaki, Y., Li, X. & Maquat, L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21, 3536–3545 (2002).
Maquat, L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5, 89–99 (2004).
Ohno, M., Segref, A., Bachi, A., Wilm, M. & Mattaj, I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187–198 (2000).
Segref, A., Mattaj, I.W. & Ohno, M. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7, 351–360 (2001).
Flaherty, S.M., Fortes, P., Izaurralde, E., Mattaj, I.W. & Gilmartin, G.M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 94, 11893–11898 (1997).
Balatsos, N.A., Nilsson, P., Mazza, C., Cusack, S. & Virtanen, A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J. Biol. Chem. 281, 4517–4522 (2006).
Fortes, P. et al. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6, 191–196 (2000).
Izaurralde, E. et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78, 657–668 (1994).
Izaurralde, E., McGuigan, C. & Mattaj, I.W. Nuclear localization of a cap-binding protein complex. Cold Spring Harb. Symp. Quant. Biol. 60, 669–675 (1995).
Görlich, D. et al. Importin provides a link between nuclear protein import and U snRNA export. Cell 87, 21–32 (1996).
Li, H. & Tschudi, C. Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol. Cell. Biol. 25, 2216–2226 (2005).
Oeffinger, M. et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4, 951–956 (2007).
Wen, Y. & Shatkin, A.J. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-α. Genes Dev. 14, 2944–2949 (2000).
Görlich, D., Panté, N., Kutay, U., Aebi, U. & Bischoff, F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594 (1996).
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14, 229–233 (2007).
Conti, E. & Kuriyan, J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure 8, 329–338 (2000).
Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6, 388–397 (1999).
Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 (1998).
Fontes, M.R., Teh, T. & Kobe, B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 (2000).
Fontes, M.R., Teh, T., Jans, D., Brinkworth, R.I. & Kobe, B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-α. J. Biol. Chem. 278, 27981–27987 (2003).
Mazza, C., Ohno, M., Segref, A., Mattaj, I.W. & Cusack, S. Crystal structure of the human nuclear cap binding complex. Mol. Cell 8, 383–396 (2001).
Mazza, C., Segref, A., Mattaj, I.W. & Cusack, S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21, 5548–5557 (2002).
Calero, G. et al. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 9, 912–917 (2002).
Olia, A.S. et al. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol. 363, 558–576 (2006).
Worch, R. et al. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA 11, 1355–1363 (2005).
Koch, M.H., Vachette, P. & Svergun, D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 36, 147–227 (2003).
Putnam, C.D., Hammel, M., Hura, G.L. & Tainer, J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007).
Petoukhov, M.V. & Svergun, D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 (2005).
Cingolani, G., Petosa, C., Weis, K. & Muller, C.W. Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 (1999).
Svergun, D.I., Barberato, C. & Koch, M.H.J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995).
García De La Torre, J., Huertas, M.L. & Carrasco, B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78, 719–730 (2000).
Wilson, K.F. et al. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J. Biol. Chem. 274, 4166–4173 (1999).
Wilson, K.F., Wu, W.J. & Cerione, R.A. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J. Biol. Chem. 275, 37307–37310 (2000).
Mamane, Y. et al. eIF4E-from translation to transformation. Oncogene 23, 3172–3179 (2004).
Bischoff, F.R. & Ponstingl, H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82 (1991).
Coutavas, E., Ren, M., Oppenheim, J.D., D Eustachio, P. & Rush, M.G. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature 366, 585–587 (1993).
Bischoff, F.R., Krebber, H., Smirnova, E., Dong, W. & Ponstingl, H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14, 705–715 (1995).
Lührmann, R., Kastner, B. & Bach, M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087, 265–292 (1990).
Izaurralde, E. & Mattaj, I.W. Transport of RNA between nucleus and cytoplasm. Semin. Cell Biol. 3, 279–288 (1992).
Svergun, D.I. Mathematical methods in small-angle scattering data analysis. J. Appl. Crystallogr. 24, 485–492 (1991).
Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Leslie, A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 (1992).
Evans, P.R. in Proceedings of CCP4 Study Weekend: Data Collection and Processing (eds. Sawyer, L., Isaacs, N. & Bailey, S.) Science and Engineering Research Council, 114–122 (Daresbury Laboratory, Warrington, UK, 1993).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Konarev, P.V., Volkov, V.V., Sokolova, A.V., Koch, M.H.J. & Svergun, D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003).
Semisotnov, G.V., Timchenko, A.A., Melnik, B.S., Kimura, K. & Kihara, H. Kratky plot as a tool to evaluate the molecular mass of globular proteins. Photon Factory Activity Report 2002 20, Part B 256 (2003).
Laue, T.M., Shah, B.D., Ridgeway, T.M. & Pelletier, S.L. Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds. Harding, S.E., Rowe, A.J., & Horton, J.C.) 90–125 (Royal Society of Chemistry, Cambridge, UK, 1992).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant number GM40654. We would like to thank G. Cingolani (State University of New York Upstate Medical University) for assistance with the agarose native gel assays and discussion of the results and for the importin-β constructs encoding residues 1–619, 1–445, 45–876 (pTYB4 vector (NEB)) and 127–876 (pQE60 (Qiagen)); H. Sondermann and Q. Wang for helping with the Advanced Photon Source (APS) data collection; B. Crane, M. Sanches and A. Rojas for helpful discussions; and C. Westmiller for her excellent secretarial assistance. We also thank the Cornell High Energy Synchrotron Source (CHESS) and APS beamline staff for their assistance with the data collection.
Author information
Authors and Affiliations
Contributions
S.M.G.D. designed and performed the protein purifications, EMSA, crystallography, UV cross-linking assays, pull-downs, native gels, SAXS, bioinformatics work and AUC experiments, analyzed data and wrote the paper; K.F.W. designed and performed cell biology experiments, analyzed data and wrote the paper; K.S.R. performed cell biology experiments; A.L.B.A. analyzed crystallographic data; R.A.C. analyzed data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Data and Supplementary Methods (PDF 1069 kb)
Rights and permissions
About this article
Cite this article
Dias, S., Wilson, K., Rojas, K. et al. The molecular basis for the regulation of the cap-binding complex by the importins. Nat Struct Mol Biol 16, 930–937 (2009). https://doi.org/10.1038/nsmb.1649
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1649
This article is cited by
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells
Genome Biology (2024)
-
Maturation and shuttling of the yeast telomerase RNP: assembling something new using recycled parts
Current Genetics (2022)
-
The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration
Cellular and Molecular Life Sciences (2019)
-
Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses
Nature Communications (2018)
-
Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1
Nature Communications (2017)