Abstract
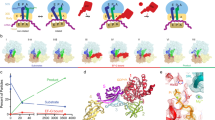
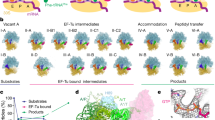
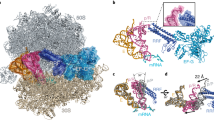
EF4 (LepA) is an almost universally conserved translational GTPase in eubacteria. It seems to be essential under environmental stress conditions and has previously been shown to back-translocate the tRNAs on the ribosome, thereby reverting the canonical translocation reaction. In the current work, EF4 was directly visualized in the process of back-translocating tRNAs by single-particle cryo-EM. Using flexible fitting methods, we built a model of ribosome-bound EF4 based on the cryo-EM map and a recently published unbound EF4 X-ray structure. The cryo-EM map establishes EF4 as a noncanonical elongation factor that interacts not only with the elongating ribosome, but also with the back-translocated tRNA in the A-site region, which is present in a previously unseen, intermediate state and deviates markedly from the position of a canonical A-tRNA. Our results, therefore, provide insight into the underlying structural principles governing back-translocation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Caldon, C.E., Yoong, P. & March, P.E. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41, 289–297 (2001).
Margus, T., Remm, M. & Tenson, T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics 8, 15 (2007).
Qin, Y. et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127, 721–733 (2006).
Colca, J.R. et al. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278, 21972–21979 (2003).
Wilson, D.N. & Nierhaus, K. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 42, 187–219 (2007).
Shoji, S., Walker, S. & Fredrick, K. Reverse translocation of tRNA in the ribosome. Mol. Cell 24, 931–942 (2006).
Konevega, A.L. et al. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat. Struct. Mol. Biol. 14, 318–324 (2007).
Frank, J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies (Academic Press, New York, 1996).
Stark, H. et al. Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell 88, 19–28 (1997).
Agrawal, R.K. et al. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J. Cell Biol. 150, 447–460 (2000).
Berk, V., Zhang, W., Pai, R. & Cate, J. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 103, 15830–15834 (2006).
Evans, R.N., Blaha, G., Bailey, S. & Steitz, T. The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. USA 105, 4673–4678 (2008).
Yusupov, M.M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Wriggers, W., Milligan, R.A. & McCammon, J.A. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999).
Sali, A. & Blundell, T. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Connell, S. et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell 25, 751–764 (2007).
Laurberg, M. et al. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 303, 593–603 (2000).
Topf, M., Baker, M., John, B., Chiu, W. & Sali, A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J. Struct. Biol. 149, 191–203 (2005).
Topf, M. et al. Protein structure fitting and refinement guided by cryoEM density. Structure 16, 295–307 (2008).
Frank, J. & Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000).
Valle, M. et al. Locking and unlocking of ribosomal motions. Cell 114, 123–134 (2003).
Nierhaus, K.H. Protein synthesis. An elongation factor turn-on. Nature 379, 491–492 (1996).
Wilson, K.S. & Noller, H.F. Molecular movement inside the translational engine. Cell 92, 337–349 (1998).
Savelsbergh, A. et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell 11, 1517–1523 (2003).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Valle, M. et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 10, 899–906 (2003).
Sanbonmatsu, K.Y., Joseph, S. & Tung, C. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. USA 102, 15854–15859 (2005).
Leach, K.L. et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol. Cell 26, 393–402 (2007).
Nyborg, J. et al. Structure of the ternary complex of EF-Tu: macromolecular mimicry in translation. Trends Biochem. Sci. 21, 81–82 (1996).
Marquez, V., Wilson, D.N., Tate, W.P., Triana-Alonso, F. & Nierhaus, K.H. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 118, 45–55 (2004).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Penczek, P.A., Frank, J. & Spahn, C. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 154, 184–194 (2006).
Acknowledgements
We thank M. Pech for his help and discussions, and A. Sali and W. Chiu for their support in the development of the cryo-EM density fitting methods. The present work was supported by grants from the Volkswagen Stiftung (C.M.T.S.), the Deutsche Forschungsgemeinschaft (DFG; SF8740 and SP 1130/2-1 to C.M.T.S. and K.H.N.), by the European Union 3D-EM Network of Excellence and by the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum). S.R.C. was supported with a grant from the Alexander von Humboldt Stiftung. M.T. is supported by a Medical Research Council Career Development Award (G0600084).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–4, Supplementary Results and Supplementary Methods. (PDF 6212 kb)
Supplementary Movie
Resolution of the switch 1 region in the cryo-EM reconstruction. The switch 1 region of EF4 (red) has been modeled according to the corresponding region seen in the X-ray structure of EF-G-2 (PDB 1WDT). The cryo-EM density map is illustrated as a wire mesh and has been rendered at two thresholds. The switch 1 region (as modeled on the X-ray structure) does not directly correspond to the density and therefore it is presumed that it undergoes a conformational change on the ribosome. The cryo-EM density suggests that this conformational change would lead to the distal end of the loop forming an interaction near helix 14 (orange) of the 16S rRNA. The helical region in the distal end of the switch region appears to interact with domain III (of EF4; cyan) and H95 (purple) of the 23S rRNA. The movie was prepared with PyMOL. (MOV 4282 kb)
Supplementary Movie 2
A 360 degree rotation around the 3' CCA end of the A/L-tRNA (blue). The cryo-EM density is shown as a wire mesh and rendered at the relatively high 3 sigma threshold such that the various elements remain well defined although somewhat fragmented. The gap between the H71 and 92 (the H71/92 corridor) is clearly occupied by density, and it is within this density that the 3' CCA end of the A/L-tRNA has been modeled. (MOV 1675 kb)
Rights and permissions
About this article
Cite this article
Connell, S., Topf, M., Qin, Y. et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol 15, 910–915 (2008). https://doi.org/10.1038/nsmb.1469
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1469
This article is cited by
-
In silico analysis of bacterial translation factors reveal distinct translation event specific pI values
BMC Genomics (2021)
-
EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nature Structural & Molecular Biology (2016)
-
West syndrome caused by homozygous variant in the evolutionary conserved gene encoding the mitochondrial elongation factor GUF1
European Journal of Human Genetics (2016)
-
The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life
BMC Genomics (2015)
-
Identification of Two Structural Elements Important for Ribosome-Dependent GTPase Activity of Elongation Factor 4 (EF4/LepA)
Scientific Reports (2015)