Abstract
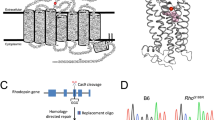
The retinitis pigmentosa 2 (RP2) gene is responsible for a particular variant of X chromosome–linked eye disease. Previously, RP2 was shown to bind the GTP form of the small G protein Arf-like 3 (Arl3), thus qualifying as an effector. Here we present the Arl3–GppNHp–RP2 complex structure, which shows features resembling complexes with GTPase-activating proteins (GAPs). Biochemical analysis showing a 90,000-fold stimulation of the GTPase reaction together with the structure of an Arl3–GDP–AlF4−–RP2 transition state complex showed that RP2 is an efficient GAP for Arl3, with structural features similar to other GAPs. Furthermore, the effect of mutations in patients with retinitis pigmentosa correlated with their effect on catalysis, in particular the mutation of the arginine finger of RP2. The cognate G protein–GAP pair is conserved in yeast as Cin4–Cin2, and the ability of RP2 to act as a GAP can be correlated with its ability to complement a CIN2-deletion phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Haim, M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol. Scand. Suppl. 80, 1–34 (2002).
Kennan, A., Aherne, A. & Humphries, P. Light in retinitis pigmentosa. Trends Genet. 21, 103–110 (2005).
Hims, M.M., Diager, S.P. & Inglehearn, C.F. Retinitis pigmentosa: genes, proteins and prospects. Dev. Ophthalmol. 37, 109–125 (2003).
Heckenlively, J.R., Boughman, J.A. & Friedman, L.H. Retinitis pigmentosa. (ed. Heckenlively, J.R.) 6–24 (Lippincott, Philadelphia, 1988).
Miano, M.G. et al. Identification of novel RP2 mutations in a subset of X-linked retinitis pigmentosa families and prediction of new domains. Hum. Mutat. 18, 109–119 (2001).
Schwahn, U. et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19, 327–332 (1998).
Kuhnel, K., Veltel, S., Schlichting, I. & Wittinghofer, A. Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure 14, 367–378 (2006).
Bartolini, F. et al. Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J. Biol. Chem. 277, 14629–14634 (2002).
Chapple, J.P. et al. Mutations in the N-terminus of the X-linked retinitis pigmentosa protein RP2 interfere with the normal targeting of the protein to the plasma membrane. Hum. Mol. Genet. 9, 1919–1926 (2000).
Rosenberg, T., Schwahn, U., Feil, S. & Berger, W. Genotype-phenotype correlation in X-linked retinitis pigmentosa 2 (RP2). Ophthalmic Genet. 20, 161–172 (1999).
Tian, G., Bhamidipati, A., Cowan, N.J. & Lewis, S.A. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the α/β-tubulin heterodimer. J. Biol. Chem. 274, 24054–24058 (1999).
Tian, G. et al. Pathway leading to correctly folded β-tubulin. Cell 86, 287–296 (1996).
Hoyt, M.A., Stearns, T. & Botstein, D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 10, 223–234 (1990).
Radcliffe, P.A., Hirata, D., Vardy, L. & Toda, T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol. Biol. Cell 10, 2987–3001 (1999).
Vetter, I.R. & Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001).
Linari, M., Hanzal-Bayer, M. & Becker, J. The δ subunit of rod specific cyclic GMP phosphodiesterase, PDEδ, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett. 458, 55–59 (1999).
Hanzal-Bayer, M., Renault, L., Roversi, P., Wittinghofer, A. & Hillig, R.C. The complex of Arl2-GTP and PDEδ: from structure to function. EMBO J. 21, 2095–2106 (2002).
Burd, C.G., Strochlic, T.I. & Gangi, S. Sr. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 14, 687–694 (2004).
Avidor-Reiss, T. et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527–539 (2004).
Cuvillier, A. et al. LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci. 113, 2065–2074 (2000).
Grayson, C. et al. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum. Mol. Genet. 11, 3065–3074 (2002).
Schrick, J.J., Vogel, P., Abuin, A., Hampton, B. & Rice, D.S. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am. J. Pathol. 168, 1288–1298 (2006).
Kobayashi, A., Kubota, S., Mori, N., McLaren, M.J. & Inana, G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 534, 26–32 (2003).
Linari, M. et al. The retinitis pigmentosa GTPase regulator, RPGR, interacts with the δ subunit of rod cyclic GMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 96, 1315–1320 (1999).
Meindl, A. et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat. Genet. 13, 35–42 (1996).
Kobayashi, A. et al. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest. Ophthalmol. Vis. Sci. 41, 3268–3277 (2000).
Bos, J.L., Rehmann, H. & Wittinghofer, A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 (2007).
Bowzard, J.B., Cheng, D., Peng, J. & Kahn, R.A. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282, 17568–17580 (2007).
Scheffzek, K. et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997).
Rittinger, K., Walker, P.A., Eccleston, J.F., Smerdon, S.J. & Gamblin, S.J. Structure at 1.65 of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature 389, 758–762 (1997).
Nassar, N., Hoffman, G.R., Manor, D., Clardy, J.C. & Cerione, R.A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 5, 1047–1052 (1998).
Daumke, O., Weyand, M., Chakrabarti, P.P., Vetter, I.R. & Wittinghofer, A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429, 197–201 (2004).
Pan, X., Eathiraj, S., Munson, M. & Lambright, D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303–306 (2006).
Bhamidipati, A., Lewis, S.A. & Cowan, N.J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096 (2000).
Stearns, T., Hoyt, M.A. & Botstein, D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics 124, 251–262 (1990).
Hoyt, M.A., Macke, J.P., Roberts, B.T. & Geiser, J.R. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics 146, 849–857 (1997).
Jacobs, C.W., Adams, A.E., Szaniszlo, P.J. & Pringle, J.R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107, 1409–1426 (1988).
Pasqualato, S., Renault, L. & Cherfils, J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for 'front-back' communication. EMBO Rep. 3, 1035–1041 (2002).
Goldberg, J. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96, 893–902 (1999).
Randazzo, P.A. & Hirsch, D.S. Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell. Signal. 16, 401–413 (2004).
Nie, Z. & Randazzo, P.A. Arf GAPs and membrane traffic. J. Cell Sci. 119, 1203–1211 (2006).
Eugster, A., Frigerio, G., Dale, M. & Duden, R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19, 3905–3917 (2000).
Bigay, J., Gounon, P., Robineau, S. & Antonny, B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature 426, 563–566 (2003).
Luo, R. et al. Mutational analysis of the Arf1*GTP/Arf GAP interface reveals an Arf1 mutant that selectively affects the Arf GAP ASAP1. Curr. Biol. 15, 2164–2169 (2005).
Mandiyan, V., Andreev, J., Schlessinger, J. & Hubbard, S.R. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 18, 6890–6898 (1999).
Bi, X., Corpina, R.A. & Goldberg, J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277 (2002).
Massof, R.W., Dagnelie, G., Benzschawel, T., Palmer, R.W. & Finkelstein, D. 1st order dynamics of visual-field loss in retinitis-pigmentosa. Clin. Vis. Sci. 5, 1–26 (1990).
Fan, Y. et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 36, 989–993 (2004).
Li, J.B. et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552 (2004).
Norton, A.W. et al. Evaluation of the 17-kDa prenyl-binding protein as a regulatory protein for phototransduction in retinal photoreceptors. J. Biol. Chem. 280, 1248–1256 (2005).
Zhang, H. et al. Deletion of PrBP/δ impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl. Acad. Sci. USA 104, 8857–8862 (2007).
Lin, W.D. et al. Identification of a polymorphism (D168N) in the XRP2 gene in Chinese. Hum. Mutat. 17, 354 (2001).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
We thank D. Kühlmann and C. Körner for technical assistance, M. Farkasovsky for support in yeast genetic experiments, I. Vetter and M. Weyand for crystallographic assistance and T. Barends, W. Blankenfeldt, Z. Guo and T. Meinhart for X-ray data collection. We are grateful to the staff of beamline X10SA at the Swiss Light Source for their support. We additionally thank W. Berger, Universität Zürich, for providing the RP2 clone, K. Siegers, Max-Planck-Institut für Biochemie, Martinsried, Germany, for providing the CIN2 deletion yeast strain and the CIN2 and CIN4 clones, and N. Cowan, New York University Medical Center, for providing the CoC clone. R.G. was supported by the Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Contributions
S.V. performed all the experiments and their analysis and contributed to their design; R.G. strongly supported crystallization and structure determination; E.E. established the yeast complementation assay and performed preliminary complementation experiments; A.W. supervised all the work, advised on the design of the experiments and wrote the manuscript with S.V.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 180 kb)
Rights and permissions
About this article
Cite this article
Veltel, S., Gasper, R., Eisenacher, E. et al. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol 15, 373–380 (2008). https://doi.org/10.1038/nsmb.1396
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1396
This article is cited by
-
A multivesicular body-like organelle mediates stimulus-regulated trafficking of olfactory ciliary transduction proteins
Nature Communications (2022)
-
Expression patterns of ciliopathy genes ARL3 and CEP120 reveal roles in multisystem development
BMC Developmental Biology (2020)
-
The psychosocial and service delivery impact of genomic testing for inherited retinal dystrophies
Journal of Community Genetics (2019)
-
Modulation of small GTPase activity by NME proteins
Laboratory Investigation (2018)
-
A novel mutation in RDH5 gene causes retinitis pigmentosa in consanguineous Pakistani family
Genes & Genomics (2018)