Abstract
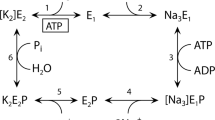
The Na,K-ATPase hydrolyzes ATP to drive the coupled extrusion and uptake of Na+ and K+ ions across the plasma membrane. Here, we report two high-resolution NMR structures of the 213-residue nucleotide-binding domain of rat α1 Na,K-ATPase, determined in the absence and the presence of ATP. The nucleotide binds in the anti conformation and shows a relative paucity of interactions with the protein, reflecting the low-affinity ATP-binding state. Binding of ATP induces substantial conformational changes in the binding pocket and in residues located in the hinge region connecting the N- and P-domains. Structural comparison with the Ca-ATPase stabilized by the inhibitor thapsigargin, E2(TG), and the model of the H-ATPase in the E1 form suggests that the observed changes may trigger the series of events necessary for the release of the K+ ions and/or disengagement of the A-domain, leading to the eventual transfer of the γ-phosphate group to the invariant Asp369.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Skou, J.C. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 23, 394–401 (1957).
Horisberger, J.D., Lemas, V., Krähenbühl, J.P. & Rossier, B.C. Structure-function relationship of Na,K-ATPase. Annu. Rev. Physiol. 53, 565–584 (1991).
Post, R.L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972).
Jorgensen, J.R. & Pedersen, P.A. Role of phylogenetically conserved amino acids in folding of Na,K-ATPase. Biochemistry 40, 7301–7308 (2001).
Kaplan, J.H. Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 (2002).
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000).
Toyoshima, C. & Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 (2002).
Zhang, P., Toyoshima, C., Yonekura, K., Green, N.M. & Stokes, D.L. Structure of the calcium pump from sarcoplasmic reticulum at 8-Å resolution. Nature 392, 835–839 (1998).
Xu, C., Rice, W.J., He, W. & Stokes, D.L. A structural model for the catalytic cycle of Ca2+-ATPase. J. Mol. Biol. 316, 201–211 (2002).
Rice, W.J., Young, H.S., Martin, D.W., Sachs, J.R. & Stokes, D.L. Structure of Na+,K+-ATPase at 11-Å resolution: comparison with Ca2+- ATPase in E1 and E2 states. Biophys. J. 80, 2187–2197 (2001).
Hebert, H., Purhonen, P., Vorum, H., Thomsen, K. & Maunsbach, A.B. Three-dimensional structure of renal Na,K-ATPase from cryo-electron microscopy of two-dimensional crystals. J. Mol. Biol. 314, 479–494 (2001).
Auer, M., Scarborough, G.A. & Kühlbrandt, W. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature 392, 840–843 (1998).
Toyofuku, T., Kurzydlowski, K., Tada, M. & MacLennan, D.H. Amino acids Lys-Asp-Asp-Lys-Pro-Val402 in the Ca2+-ATPase of cardiac sarcoplasmic reticulum are critical for functional association with phospholamban. J. Biol. Chem. 269, 22929–22932 (1994).
Jorgensen, P.L. & Pedersen, P.A. Structure-function relationships of Na+, K+, ATP, or Mg2+ binding and energy transduction in Na,K-ATPase. Biochim. Biophys. Acta 1505, 57–74 (2001).
McIntosh, D.B., Woolley, D.G., Vilsen, B. & Andersen, J.P. Mutagenesis of segment 487Phe-Ser-Arg-Asp-Arg-Lys492 of sarcoplasmic reticulum Ca2+-ATPase produces pumps defective in ATP binding. J. Biol. Chem. 271, 25778–25789 (1996).
Wang, K. & Farley, R.A. Lysine 480 is not an essential residue for ATP-binding or hydrolysis by Na,K-ATPase. J. Biol. Chem. 267, 3577–3580 (1992).
Jacobsen, M.D., Pedersen, P.A. & Jorgensen, P.L. Importance of Na,K-ATPase residue α1-Arg544 in the segment Arg544- Asp567 for high-affinity binding of ATP, ADP, or MgATP. Biochemistry 41, 1451–1456 (2002).
Teramachi, S., Imagawa, T., Kaya, S. & Taniguchi, K. Replacement of several single amino acid side chains exposed to the inside of the ATP-binding pocket induces different extents of affinity change in the high and low affinity ATP-binding sites of rat Na/K-ATPase. J. Biol. Chem. 270, 37394–37400 (2002).
Kühlbrandt, W., Zeelen, J. & Dietrich, J. Structure, mechanism, and regulation of the Neurospora plasma membrane H+-ATPase. Science 297, 1692–1696 (2002).
Jordan, C., Püschel, B., Koob, R. & Drenckhahn, D. Identification of a binding motif for ankyrin on the α-subunit of Na+,K+-ATPase. J. Biol. Chem. 270, 29971–29975 (1995).
Therien, A.G. & Blostein, R. Mechanisms of sodium pump regulation. Am. J. Physiol. Cell Physiol. 279, C541–566 (2000).
Doné, S.C. et al. Tyrosine 537 within the Na+,K+-ATPase α-subunit is essential for AP-2 binding and clathrin-dependent endocytosis. J. Biol. Chem. 277, 17108–17111 (2002).
Neri, D., Szyperski, T., Otting, G., Senn, H. & Wüthrich, K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28, 7510–7516 (1989).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Bartels, C., Xia, T.H., Billeter, M., Güntert, P. & Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 (1995).
Herrmann, T., Güntert, P. & Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002).
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Cornell, W.D. et al. A second generation force-field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995).
Koradi, R., Billeter, M. & Güntert, P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 124, 139–147 (2000).
Vriend, G. WHATIF: a molecular modeling and drug design program. J. Mol. Graph. 52, 29–36 (1990).
Laskowski, R.A., Rullmann, J.A., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Nicholls, A., Sharp, K. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Humphrey, W., Dalke, A. & Schulten, K. VMD—visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Wriggers, W., Milligan, R.A. & McCammon, J.A. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999).
Kleywegt, G.J. & Jones, T.A. A super position. ESF/CCP4 Newslett. 31, 9–14 (1994).
Garrett, D.S., Seok, Y.J., Peterkofsky, A., Clore, G.M. & Gronenborn, A.M. Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry 36, 4393–4398 (1997).
Acknowledgements
We thank J.L. van der Plas, J. Plaisier and N. Pannu for setting up software and writing several useful programs, W.J. Rice and co-workers for kindly providing the 11-Å EM map of Na,K-ATPase, R.A.G. de Graaff for critical reading of the manuscript and S. Hilge for professional help with the figures. This work was supported by grants to M.H. of the Swiss National Foundation, the Netherlands Organisation for Scientific Research (NWO) and Technologie stichting STW. G.S. acknowledges the Dutch Royal Academy of Sciences for fellowship support. P.G. acknowledges the Tatsuo Miyazawa Memorial Program of RIKEN for support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hilge, M., Siegal, G., Vuister, G. et al. ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase. Nat Struct Mol Biol 10, 468–474 (2003). https://doi.org/10.1038/nsb924
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb924
This article is cited by
-
The Na,K-ATPase acts upstream of phosphoinositide PI(4,5)P2 facilitating unconventional secretion of Fibroblast Growth Factor 2
Communications Biology (2020)
-
ANS Interacts with the Ca2+-ATPase Nucleotide Binding Site
Journal of Fluorescence (2020)
-
Exploiting image registration for automated resonance assignment in NMR
Journal of Biomolecular NMR (2015)
-
Mapping the ATP Binding Site in the Plasma Membrane H+-ATPase from Kluyveromyces lactis
Journal of Fluorescence (2014)
-
The second sodium pump: from the function to the gene
Pflügers Archiv - European Journal of Physiology (2012)