Abstract
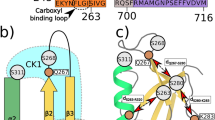
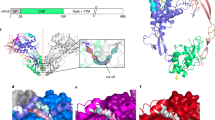
The DEP domain of Dishevelled (Dvl) proteins transduces signals to effector proteins downstream of Dvl in the Wnt pathway. Here we report that DEP-containing mutants inhibit Wnt-induced, but not Dvl-induced, activation of the transcription factor Lef-1. This inhibitory effect is weakened by a K434M mutation. Nuclear magnetic resonance spectroscopy revealed that the DEP domain of mouse Dvl1 comprises a three-helix bundle, a β-hairpin `arm' and two short β-strands at the C-terminal region. Lys 434 is located at the tip of the β-hairpin `arm'. Based on our findings, we conclude that DEP interacts with regulators upstream of Dvl via a strong electric dipole on the molecule's surface created by Lys 434, Asp 445 and Asp 448; the electric dipole and the putative membrane binding site are at two different locations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Brown, J.D. & Moon, R.T. Wnt signaling: why is everything so negative? Curr. Opin. Cell Biol. 10, 182–187 (1998).
Dale, T.C. Signal transduction by the Wnt family of ligands. Biochem. J. 329, 209–223 (1998).
Dierick, H. & Bejsovec, A. Cellular mechanisms of wingless/Wnt signal transduction. Curr. Top. Dev. Biol. 43, 153–190 (1999).
Nusse, R. WNT targets. Repression and activation. Trends Genet. 15, 1–3 (1999).
Wodarz, A. & Nusse, R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14, 59–88 (1998).
Tetsu, O. & McCormick, F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999).
He H.C. et al. Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 (1998).
Wieschaus, E., Nusslein-Volhard, C. & Kluding, H. Kruppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev. Biol. 104, 172–186 (1984).
Yanagawa, S., van Leeuwen, F., Wodarz, A., Klingensmith, J. & Nusse, R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes Dev. 9, 1087–1097 (1995).
Li, L. et al. Dishevelled proteins lead to two signaling pathways. Regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J. Biol. Chem. 274, 129–134 (1999).
Tomlinson, A., Strapps, W.R. & Heemskerk, J. Linking Frizzled and Wnt signaling in Drosophila development. Development 124, 4515–4521 (1997).
Park, M., Wu, X., Golden, K., Axelrod, J.D. & Bodmer, R. The wingless signaling pathway is directly involved in Drosophila heart development. Dev. Biol. 177, 104–116 (1996).
Farr, G.H., III et al. Interaction among GSK-3, GBP, axin, and APC in Xenopus axis specification. J. Cell Biol. 148, 691–702 (2000).
Ikeda, S. et al. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta- dependent phosphorylation of beta-catenin. EMBO J. 17, 1371–1384 (1998).
Itoh, K., Antipova, A., Ratcliffe, M.J. & Sokol, S. Interaction of Dishevelled and Xenopus axin-related protein is required for Wnt signal transduction. Mol. Cell Biol. 20, 2228–2238 (2000).
Li, L. et al. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt- mediated regulation of LEF-1. EMBO J. 18, 4233–4240 (1999).
Smalley, M.J. et al. Interaction of axin and dvl-2 proteins regulates dvl-2-stimulated TCF-dependent transcription. EMBO J. 18, 2823–2835 (1999).
Yuan, H., Mao, J., Li, L. & Wu, D. Suppression of glycogen synthase kinase activity is not sufficient for leukemia enhancer factor-1 activation. J. Biol. Chem. 274, 30419–30423 (1999).
Theisen, H. et al. Dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development 120, 347–360 (1994).
Heisenberg, C.P. et al. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405, 76–81 (2000).
Wallingford, J.B. et al. Dishevelled controls cell polarity during Xenopus gastrulation. Nature 405, 81–85 (2000).
Boutros, M., Paricio, N., Strutt, D.I. & Mlodzik, M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94, 109–118 (1998).
Boutros, M. & Mlodzik, M. Dishevelled: at the crossroads of divergent intracellular signaling pathways. Mech. Dev. 83, 27–37 (1999).
Ponting, C.P. & Bork, P. Pleckstrin's repeat performance: a novel domain in G-protein signaling? Trends Biochem. Sci. 21, 245–246 (1996).
Kharrat, A. et al. Conformational stability studies of the pleckstrin DEP domain: definition of the domain boundaries. Biochim. Biophys. Acta 1385, 157–164 (1998).
Zheng, J. et al. The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains. J. Biol. Chem. 272, 30340–30344 (1997).
Zheng, J. et al. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J. Mol. Biol. 255, 14–21 (1996).
Hsu, S.C., Galceran, J. & Grosschedl, R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell Biol. 18, 4807–4818 (1998).
Sakanaka, C., Weiss, J.B. & Williams, L.T. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc. Natl. Acad. Sci. USA 95, 3020–3023 (1998).
Fushman, D. et al. The solution structure and dynamics of the pleckstrin homology domain of G protein-coupled receptor kinase 2 (beta-adrenergic receptor kinase 1). A binding partner of Gbetagamma subunits. J. Biol. Chem. 273, 2835–2843 (1998).
Guntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Rothbacher, U. et al. Dishevelled phosphorylation, subcellular localization and multimerization regulate its role in early embryogenesis. EMBO J. 19, 1010–1022 (2000).
Axelrod, J.D., Miller, J.R., Shulman, J.M., Moon, R.T. & Perrimon, N. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev. 12, 2610–2622 (1998).
Koelle, M.R. & Horvitz, H.R. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84, 115–125 (1996).
Murray, D. et al. Electrostatic properties of membranes containing acidic lipids and adsorbed basic peptides: theory and experiment. Biophys. J. 77, 3176–3188 (1999).
Gelb, M.H., Cho, W. & Wilton, D.C. Interfacial binding of secreted phospholipases A(2): more than electrostatics and a major role for tryptophan. Curr. Opin. Struct. Biol. 9, 428–432 (1999).
Sheinerman, F.B., Norel, R. & Honig, B. Electrostatic aspects of protein–protein interactions. Curr. Opin. Struct. Biol. 10, 153–159 (2000).
Honig, B. & Nicholls, A. Classical electrostatics in biology and chemistry. Science 268, 1144–1149 (1995).
Semenov, M.V. & Snyder, M. Human dishevelled genes constitute a DHR-containing multigene family. Genomics 42, 302–310 (1997).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Ramakrishnan, V., Finch, J.T., Graziano, V., Lee, P.L. & Sweet, R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223 (1993).
Belikov, S. & Karpov, V. Linker histones: paradigm lost but questions remain. FEBS Lett. 441, 161–164 (1998).
Krasnow, R.E., Wong, L.L. & Adler, P.N. Dishevelled is a component of the frizzled signaling pathway in Drosophila. Development 121, 4095–4102 (1995).
Clore, G.M. & Gronenborn, A.M. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 239, 349–363 (1994).
Matsuo, H., Kupce, E., Li, H. & Wagner, G. Increased sensitivity in HNCA and HN(CO)CA experiments by selective C beta decoupling. J. Magn. Reson. B 113, 91–96 (1996).
Kay, L.E. & Bax, A. New methods for the measurement of NH-CαH coupling constants in 15N-labeled proteins. J. Magn. Res. 86, 110–126 (1990).
Xia, T.-H., Bartels, C. & Wüthrich, K. XEASY, ETH automated spectroscopy for X window system, user mannal (ETH-Honggerberg, Zurich; 1993).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–32 (1996).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Bacon, D.J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
This work was funded by the American Lebanese Syrian Associated Charities to, by a Cancer Center (CORE) support grant from the National Cancer Institute and by a research grant from the National Institute of General Medical Science. We thank S. White and D. Cowburn for critical reading and comments on the manuscript; P. Mehta for help with data base searching and protein sequence analysis; J. Cay Jones, A. McArthur, C. Ross, and F. Witte for scientific editing. J.T.N is a trainee of the professional oncology program (POE) at St. Jude, and was funded by the National Cancer Institute and American Lebanese Syrian Associated Charities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wong, H., Mao, J., Nguyen, J. et al. Structural basis of the recognition of the Dishevelled DEP domain in the Wnt signaling pathway. Nat Struct Mol Biol 7, 1178–1184 (2000). https://doi.org/10.1038/82047
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/82047
This article is cited by
-
Wnt/β-catenin signaling pathway in uterine leiomyoma: role in tumor biology and targeting opportunities
Molecular and Cellular Biochemistry (2021)
-
Knockdown of DEPDC1B inhibits the development of glioblastoma
Cancer Cell International (2020)
-
Dishevelled-3 conformation dynamics analyzed by FRET-based biosensors reveals a key role of casein kinase 1
Nature Communications (2019)
-
Suppression of planar cell polarity signaling and migration in glioblastoma by Nrdp1-mediated Dvl polyubiquitination
Oncogene (2017)
-
Identification of DEP domain-containing proteins by a machine learning method and experimental analysis of their expression in human HCC tissues
Scientific Reports (2016)