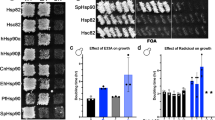
Abstract
Both randomized oligonucleotide cassette mutagenesis and site-directed mutagenesis have been used in combination with a yeast genetic screen to identify critical residues in the DNA-binding domain of heat shock transcription factor from Saccharomyces cerevisiae. Most of the surface residues in this highly conserved domain can be changed to alanine with no observable effect on function. Of nine critical residues identified in this screen, five are within helix α3, previously designated as the probable DNA recognition helix in the crystal structure of the Kluyveromyces lactis protein. The other four residues may be involved in DNA-binding or protein–protein interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lis, J. & Wu, C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell 74, 1–4 (1993).
Wiederrecht, G., Seto, D. & Parker, C.S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54, 841–853 (1988).
Flick, K.E., Gonzalez, L., Harrison, C.J. & Nelson, H.C.M. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains. J. biol. Chem. 269, 12475–12481 (1994).
Sorger, P.K. & Nelson, H.C. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell 59, 807–813 (1989).
Harrison, C.J., Bohm, A.A. & Nelson, H.C.M. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263, 224–227 (1994).
Schultz, S.C., Shields, G.C. & Steitz, T.A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253, 1001–1007 (1991).
Vuister, G.W., Kim, S.J., Wu, C. & Bax, A. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix-turn-helix and HNF-3/ forkhead families of transcription factors. Biochemistry 33, 10–6 (1994).
Xiao, H. & Lis, J.T. Germline transformation used to define key features of heat-shock response elements. Science 239, 1139–1142 (1988).
Amin, J., Ananthan, J. & Voellmy, R. Key features of heat shock regulatory elements. Molec. Cell. Biol. 8, 3761–3769 (1988).
Perisic, O., Xiao, H. & Lis, J.T. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell 59, 797–806 (1989).
Kim, S.-J., Tsukiyama, T., Lewis, M.S. & Wu, C. Interaction of DNA-bindig domain of Drosophila heat shock factor with its cognate DNA site: A thermodynamic analysis using analytical ultracentrifugation. Prot. Sci. 3, (1994).
Amin, J., Fernandez, M., Ananthan, J., Lis, J.T. & Voellmy, R. Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro. J. biol. Chem. 269, 4804–4811 (1994).
Gross, D.S., English, K.E., Collins, K.W. & Lee, S.W. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. molec. Biol. 216, 611–631 (1990).
Rye, H.S., Drees, B.L., Nelson, H.C. & Glazer, A.N. Stable fluorescent dye-DNA complexes in high sensitivity detection of protein-DNA interaction. Application to heat shock transcription factor. J. biol. Chem. 268, 25229–25238 (1993).
Liu-Johnson, H.N., Gartenberg, M.R. & Crothers, D.M. The DNA binding domain and bending angle of E. coli CAP protein. Cell 47, 995–1005 (1986).
Bonner, J.J., Heyward, S. & Fackenthal, D.L. Temperature-dependent regulation of a heterologous transcriptional activation domain fused to yeast heat shock transcription factor. Molec. Cell Biol. 12, 1021–1030 (1992).
Silar, P., Butler, G. & Thiele, D.J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Molec. Cell Biol. 11, 1232–1238 (1991).
Yang, W.M., Gahl, W. & Hamer, D. Role of heat shock transcription factor in yeast metallothionein gene expression. Molec. Cell Biol. 11, 3676–3681 (1991).
Sorger, P.K. & Pelham, H.R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54, 855–864 (1988).
Jakobsen, B.K. & Pelham, H.R. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 10, 369–375 (1991).
Jakobsen, B.K. & Pelham, H.R. Constitutive binding of yeast heat shock factor to DNA in vivo. Molec. Cell Biol. 8, 5040–5042 (1988).
Kunkel, T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. natn Acad. Sci. U.S.A. 82, 488–492 (1985).
Hill, D.E., Oliphant, A.R. & Struhl, K. Mutagenesis with degenerate oligonucleotides: an efficient method for saturating a defined DNA region with base pair substitutions. Meth. Enzymol. 155, 558–568 (1987).
Reidhaar-Olson, J.F. & Sauer, R.T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science 241, 53–57 (1988).
Bowie, J.U., Reidhaar, O.J., Lim, W.A. & Sauer, R.T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science 247, 1306–1310 (1990).
Cunningham, B.C. & Wells, J.A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 (1989).
Wells, J.A. Systematic mutational analyses of protein-protein interfaces. Meth. Enzymol. 202, 390–411 (1991).
Pabo, C.O. & Sauer, R.T. Transcription factors: structural families and principles of DNA recognition. A. Rev. Biochem. 61, 1053–1095 (1992).
Shore, D. & Nasmyth, K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51, 721–732 (1987).
Elble, R. A simple and efficient procedure for transformation of yeasts. BioTechniques 13, 18–20 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hubl, S., Owens, J. & Nelson, H. Mutational analysis of the DNA-binding domain of yeast heat shock transcription factor. Nat Struct Mol Biol 1, 615–620 (1994). https://doi.org/10.1038/nsb0994-615
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0994-615
This article is cited by
-
Some like it hot
Nature Structural & Molecular Biology (1994)