Abstract
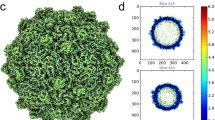
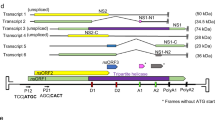
Numerous small, RNA-containing insect viruses are currently classified as picornaviruses, or as 'picorna-like', since they superficially resemble the true picornaviruses. Considerable evidence now suggests that several of these viruses are members of a distinct family. We have determined the gene sequence of the capsid proteins and the 2.4 Å resolution crystal structure of the cricket paralysis virus. While the genome sequence indicates that the insect picorna-like viruses represent a distinct lineage compared to true picornaviruses, the capsid structure demonstrates that the two groups are related. These viral genomes are, thus, best viewed as composed of exchangeable modules that have recombined.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Reinganum, C., O'Loughlin, G.T. & Hogan, T.W. A nonoccluded virus of the field crickets Teleogryllus oceanicus and T. commodus. J. Invertebr. Pathol. 16, 214–219 (1970).
Jousset, F.X., Plus, N., Croizier, G. & Thomas, M. [Existence in Drosophila of 2 groups of picornavirus with different biological and serological properties]. [French]. Comptes Rendus Hebdomadaires des Seances de l Academie des Sciences - D: Sciences Naturelles 275, 3043–3046 (1972).
Johnson, K.N. & Christian, P.D. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J. Gen. Virol. 79, 191–203 ( 1998).
Moore, N.F., Kearns, A. & Pullin, J.S.K. Characterization of cricket paralysis virus-induced polypeptides. J. Virol. 33, 1– 9 (1980).
King, L.A. & Moore, N.F. Evidence for the presence of a genome-linked protein in two insect picornaviruses, cricket paralysis and Drosophila C viruses. FEMS Microbiol. Lett. 26, 121–127 (1988).
Christian, P.D. & Scotti, P.D. Picornaviruses in insects. In The insect viruses (eds. Miller, L.K. & Ball, L.A.) (Plenum Publishing Corporation, New York; 1999 ).
Murphy, F.A., et al. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses (Springer-Verlag, New York & Vienna, 1995).
Scotti, P.D. The estimation of virus density in isopycnic cesium chloride gradients. J. Virol. Methods 12, 149–60 (1985).
Jousset, F.X., Bergoin, M. & Revet, B. Characterization of the Drosophila C virus. J. Gen. Virol. 34, 269–283 (1977).
Plus, N., Croizier, G., Reinganum, C. & Scotti, P.D. Cricket paralysis virus and Drosophila C virus: serological analysis and comparison of capsid polypeptides and host range. J. Invertebr. Pathol. 31, 296–302 ( 1978).
Scotti, P.D., Longworth, J.F., Plus, N., Croizier, G. & Reinganum, C. The biology and ecology of strains of an insect small RNA virus complex. Adv. Virus Res. 26, 117–143 (1981).
Pelletier, J. & Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 ( 1988).
Jacobson, M.F. & Baltimore, D. Polypeptide cleavages in the formation of poliovirus proteins. Proc. Natl. Acad. Sci. USA 61, 77–84 (1968).
Reavy, B. & Moore, N.F. The gene organization of a small RNA-containing insect virus: comparison with that of mammalian picornaviruses. Virology 131, 551–554 (1983).
Koonin, E.V. & Gorbalenya, A.E. An insect picornavirus may have genome organization similar to that of caliciviruses. FEBS Lett. 297, 81–86 ( 1992).
Moore, N.F., Reavy, B., Pullin, J.S.K. & Plus, N. The polypeptides induced in Drosophila cells by Drosophila C virus (strain Ouarzazate). Virology 112, 411– 416 (1981).
Sasaki, J., Nakashima, N., Saito, H. & Noda, H. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3´ part of the genome. Virology 244, 50–58 (1998).
Hogle, J.M., Chow, M. & Filman, D.J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229, 1358– 1365 (1985).
Arnold, E. et al. The structure determination of a common cold virus, human rhinovirus-14. Acta Crystallogr. A43, 346 –361 (1987).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119 (1991).
Caspar, D.L.D. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27, 1–24 ( 1962).
Rossmann, M.G. et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145 –153 (1985).
Colston, E. & Racaniello, V.R. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13, 5855–5862 (1994).
Smith, T., et al. The site of attachment in human rhino virus 14 for antiviral agents that inhibit uncoating. Science 233, 1286–1293 (1986).
Filman, D.J. et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8, 1567–1579 (1989).
Hadfield, A.T. et al. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5, 427–441 (1997).
Muckelbauer, J.K. et al. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3, 653–667 (1995).
Heinz, B., Shepard, D. & Rueckert, R. Escape mutant analysis of a drug binding site can be used to map functions in the rhinovirus capsid. In Use of X-ray crystallography in the design of anti-viral agents (eds. Laver, W. & Air, G.) 173–186 (Academic Press, San–Diego, 1990).
Kaplan, G., Freistadt, M.S. & Racaniello, V.R. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol. 64, 4697– 4702 (1990).
Burroughs, J.N., Rowlands, D.J., Sangar, D.V., Talbot, P. & Brown, F. Further evidence for multiple proteins in the foot-and-mouth disease virus particle. J. Gen. Virol. 13, 73–84 (1971).
Luo, M. et al. The atomic structure of Mengo virus at 3.0 Å resolution. Science 235, 182–191 ( 1987).
Chow, M. et al. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327, 482– 486 (1987).
Bennett, M.J., Schlunegger, M.P. & Eisenberg, D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 4, 2455–2468 (1995).
Basavappa, R. et al. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 3, 1651– 1669 (1994).
Zlotnick, A. et al. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single asparatic acid residue. J. Biol. Chem. 269, 13680–13684 (1994).
Fisher, A.J. & Johnson, J.E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361, 176–179 (1993).
Munshi, S., et al. The 2.8 Å structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 261, 1–10 (1996).
Jacobson, M.F., Asso, J. & Baltimore, D. Further evidence on the formation of poliovirus proteins. J. Mol. Biol. 49, 657– 669 (1970).
Lomonossoff, G.P. & Johnson, J.E. The synthesis and structure of comovirus capsids. Prog. Biophys. Mol. Biol. 55, 107–137 (1991).
Hendrix, R., Smith, M., Burns, R., Ford, M. & Hatfull, G. Evolutionary relationships among diverse bacteriophages and prophages: All the world's a phage. Proc. Natl. Acad. Sci. USA 96, 2192–2197 ( 1999).
Scotti, P.D., Hoefakker, P. & Dearing, S. The production of cricket paralysis virus in suspension cultures of insect cell lines. J. Invertebr. Pathol. 68, 109–112 (1996).
Otwinowski, Z. Data collection and processing. In Proceedings of the CCP4 Study Weekend: Data Collection and Processing (eds. Sawyer, L., Isaacs, N. & Baily, S.) 56–62 (Science and Engineering Research Council, Daresbury Laboratory, Daresbury, England, 1993).
Tong, L.A. & Rossmann, M.G. The locked rotation function. Acta Crystallogr. A46, 783– 792 (1990).
Brünger, A.T. X-PLOR Version 3.0 (Yale University, New Haven, Connecticut, 1992).
CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760– 763 (1994).
Jones, T.A. A set of Averaging programs. In Proceedings of the CCP4 Study Weekend: Molecular replacement (eds. Dodson, E.J., Glover, S. & Wolf, W.) 91–105 (Science and Engineering Research Council, Daresbury Laboratory, Daresbury, England, 1992).
Kleywegt, G.J. & Jones, T.A. Halloween ... Masks and Bones. In Proceedings of the CCP4 Study Weekend: From First Map to Final Model (eds. Bailey, S., Hubbard, R. & Waller, D.) 56–66 (Science and Engineering Research Council, Daresbury Laboratory, Daresbury, England, 1994).
Brünger, A.T. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472– 474 (1992).
Rice, L.M. & Brünger, A.T. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins 19, 277–290 (1994).
Kabsch, W. & Sander, D. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced coloring capacities. J. Mol. Graph. 15, 132–134 (1997).
Kraulis, P.J. MOLSCRIPT: a program to produce detailed and schematic plots of protein structure. J. Appl. Crystallogr. 24, 946– 950 (1991).
Merritt, E.A. & Murphy, M.E.P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D50, 869–873 (1994).
Acknowledgements
We would like to thank F. Weyts, K. Johnson, A. Claudianos, A.-M. Wilkes and N. Gibb for their assistance with cloning and sequencing of the CrPV genome; S. Dearing and P. Hoefakker for assistance with the production of CrPV; V. Reddy, X.F. Dong and A. Kumar for their help in data collection; H. Giesing for comments and discussions; and B. Sheehan for assistance with computing and figures for this manuscript. Data were collected on beamline F-1 of the Cornell High Energy Synchrotron Source (CHESS), with time awarded through CHESS proposal 205. This work was supported by funding from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tate, J., Liljas, L., Scotti, P. et al. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Mol Biol 6, 765–774 (1999). https://doi.org/10.1038/11543
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/11543
This article is cited by
-
Double-stranded RNA virus outer shell assembly by bona fide domain-swapping
Nature Communications (2017)
-
A Novel Strategy for Live Detection of Viral Infection in Drosophila melanogaster
Scientific Reports (2016)
-
Assembled molecular face-rotating polyhedra to transfer chirality from two to three dimensions
Nature Communications (2016)
-
Hepatitis A virus and the origins of picornaviruses
Nature (2015)
-
Structure of Ljungan virus provides insight into genome packaging of this picornavirus
Nature Communications (2015)