Abstract
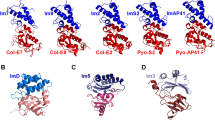
The crystal structure of the cytotoxic endonuclease domain from the bacterial toxin colicin E9 in complex with its cognate immunity protein Im9 reveals that the inhibitor does not bind at the active site, the core of which comprises the HNH motif found in intron-encoded homing endonucleases, but rather at an adjacent position leaving the active site exposed yet unable to bind DNA because of steric and electrostatic clashes with incoming substrate. Although its mode of action is unorthodox, Im9 is a remarkably effective inhibitor since it folds within milliseconds and then associates with its target endonuclease at the rate of diffusion to form an inactive complex with sub-femtomolar binding affinity. This hyperefficient mechanism of inhibition could be well suited to other toxic enzyme systems, particularly where the substrate is a polymer extending beyond the boundaries of the active site.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
James, R., Lazdunski, C. & Pattus, F (eds.) Bacteriocins, microcins and lantibiotics, NATO ASI Series H (Springer, Heidelberg; 1992).
James, R., Kleanthous, C. & Moore, G.R. The biology of E colicins: paradigms and paradoxes. Microbiology 142, 1569– 1580 (1996).
Di Masi, D.R., White, D.C., Schnaitman, C.A. & Bradbeer, C. Transport of vitamin B12 in Escherichia coli; common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J. Bacteriol. 115, 506–513 (1973).
Lazdunski, C., Bouveret, E., Rigal, A., Journet, L., Lloubès, R. & Bénédetti, H. Colicin import into Escherichia coli cells. J. Bacteriol. 180, 4993–5002 (1998).
Cramer, W.A. & Stauffacher, C.V. Structure–function of the channel-forming colicins. Annu. Rev. Biophys. Biomol. Struct . 24, 611–641 ( 1995).
Senior, B.W. & Holland, I.B. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc. Natl. Acad. Sci. USA 68, 959–963 ( 1971).
Schaller, K. & Nomura, M. Colicin E2 is a DNA endonuclease. Proc. Natl. Acad. Sci. USA 73, 3989– 3993 (1976).
Wiener, M.C., Freyman, D.M., Glosh, P. & Stroud, R.M. The crystal structure of colicin Ia. Nature 385, 461– 464 (1997).
Bénédetti, H., Lloubès, R., Lazdunski, C. & Letellier, L. Colicin A unfolds during its translocation into Escherichia coli cells and spans the whole cell envelope when its pore has formed. EMBO J. 11, 441–447 ( 1992).
Jakes, K. & Zinder, N. D. Highly purified colicin E3 contains immunity protein. Proc. Natl. Acad. Sci. USA 71, 3380–3384 (1974).
Wallis, R., Reilly, A., Rowe, A., Moore, G., James, R. & Kleanthous, C. In vivo and in vitro characterisation of overproduced colicin E9 immunity protein. Eur. J. Biochem. 207, 687– 695 (1992).
Kleanthous, C., Hemmings, A.M., Moore, G.R. & James, R. Immunity proteins and their specificity for endonuclease colicins: Telling right from wrong in protein–protein recognition. Mol. Microbiol . 28, 227–233 ( 1998).
Wallis, R., Moore, G.R., James, R. & Kleanthous, C. Protein–protein interactions in colicin E9 DNase–immunity protein complexes. Diffusion controlled association and femtomolar binding for the cognate complex. Biochemistry 34, 13743–13750 (1995).
Jackson, S.E. & Fersht, A.R. Folding of chymotrypsin inhibitor-2. Evidence for a 2-state transition. Biochemistry 30, 10428–10435 (1991).
Jackson, S.E. How do small single-domain proteins fold? Folding & Design 3, R81–R91 ( 1998).
Wallis, R., Reilly, A., Barnes, K., Abell, C., Campbell, D.G., Moore, G.R., James, R. & Kleanthous, C. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur. J. Biochem. 220, 447–454 (1994).
Garinot-Schneider, C., Pommer, A.J., Moore, G.R., Kleanthous, C. & James, R. Identification of putative active site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 260, 731–742 (1996).
Kühlmann, U.C., Kleanthous, C., James, R., Moore, G.R. & Hemmings, A.M. Preliminary crystallographic analysis of the complex between the DNase domain of colicin E9 and its cognate immunity protein. Acta Crystallogr. D 55, 256–259 (1999).
Osborne, M. J., Breeze, A. L., Lian, L-Y., Reilly, A., James, R., Kleanthous, C. & Moore, G. R. Three-dimensional solution structure and 13C NMR asignments of the colicin E9 immunity protein Im9. Biochemistry 35, 9505–9512 (1996).
Chak, K-F., Safo, M.K., Ku, W-Y, Hsieh, S-Y & Yuan, H. The crystal-structure of the immunity protein of colicin E7 suggests a possible colicin-interacting surface. Proc. Natl. Acad. Sci. U.S.A. 93, 6437–6442 (1996).
Dennis, C.A., Videler, H., Pauptit, R.A., Wallis, R., James, R., Moore, G.R. & Kleanthous, C. A structural comparison of the colicin immunity proteins Im7 and Im9 gives new insights into the molecular determinants of immunity protein specificity. Biochem. J. 333, 183–191 (1998).
Wallis, R., Leung, K-Y., Osborne, M. J., James, R., Moore, G.R. & Kleanthous, C. Specificity in protein–protein recognition: Conserved Im9 residues are the major determinants of stability in the colicin E9 DNase–Im9 complex. Biochemistry 37, 476– 485 (1998).
Li, W., Hamill, S.J., Hemmings, A.M., Moore, G.R., James, R. & Kleanthous, C. Dual recognition and the role of specificity-determining residues in colicin E9 DNase–immunity protein interactions. Biochemistry 37, 11771–11779 (1998).
Holm, L. & Sander, C. Protein-structure comparison by alignment of distance matrices J. Mol. Biol. 233, 123–138 (1993).
Janin, J. Elusive affinities. Proteins 21, 30– 39 (1995).
Osborne, M.J., Wallis, R., Leung, K-Y., William, G., Lian, L-Y., James, R., Kleanthous, C. & Moore, G.R. Identification of critical residues in the colicin E9 DNase binding region of the Im9 protein. Biochem. J. 323, 823 –831 (1997).
Li, W., Dennis, C.A., Moore, G.R., James, R. & Kleanthous, C. Protein–protein interaction specificity of Im9 for the endonuclease toxin colicin E9 defined by homologue scanning mutagenesis, J. Biol. Chem. 272, 22253–22258 (1997).
Curtis, M.D. & James, R. Investigation of the specificity of the interaction between colicin E9 and its immunity protein by site-directed mutagenesis. Mol. Microbiol. 11, 2727– 2733 (1991).
Janin, J. & Chothia, C. The structure of protein–protein recognition sites. J. Biol. Chem. 265, 16027 –16030 (1990).
Pommer, A.J., Wallis, R., Moore, R., James, R. & Kleanthous, C. Enzymological characterisation of the nuclease domain from the bacterial toxin colicin E9. Biochem. J. 334 , 387–392 (1998).
Orpen, A.G., Brammer, L., Allen, F.H., Kennard, O., Watson, D.G. & Taylor, R. Tables of bond lengths determined by X-ray and neutron diffraction. 2. Organometallic compounds and co-ordination complexes of the D-block and F-block metals. J. Chem. Soc. Dalton Trans. S1–S83 (1989).
Shub, D.A., Goodrich-Blair, H. & Eddy, S.R. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends. Biochem. Sci. 19, 402–404 (1994).
Mueller, J.E., Bryk, M., Loizos, N. & Belfort, M. in Nucleases 2nd edn (eds. Linn, S.M., Lloyd, R.S. & Roberts, R.J.) 111– 143 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; (1993).
Belfort, M. & Roberts, R.J. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25, 3379–3388 (1997).
Duan, X.Q., Gimble, F.S. & Quiocho, F.A. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell 89, 555–564 (1997).
Flick, K.E., Jurica, M.S., Monnat Jr, R.J. & Stoddard, B.L. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature 394, 96–101 (1998).
Derbyshire, V., Kowalski, J.C., Dansereau, J.T., Hauer, C.R. & Belfort, M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 265 , 494–506 (1997).
Bode, W. & Huber, R. Natural protein proteinase inhibitors and their interactions with proteinases. Eur. J. Biochem. 204, 433–451 (1992).
Guillet, V., Lapthorn, A., Hartley, R.W. & Mauguen, Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure 1, 165– 176 (1993).
Kobe, B. & Deisenhofer, J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186 (1995).
Knighton, D.R., Zheng, J.H., Teneyck, L.F., Ashford, V.A., Xuong, N.H., Taylor, S.S. & Sowadski, J.M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine-monophosphate dependent protein-kinase. Science 253 414–420 (1991).
Mol, C.D., Arvai, A.S., Sanderson, R.J., Slupphaug, G., Kavli, B., Krokan, H.E., Mosbaugh, D.W. & Tainer, J.A. Crystal-structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell 82, 701–708 ( 1995).
Kabsch, W., Mannherz, H.G., Suck, D., Pai, E.F. & Holmes, K.C. Atomic structure of the actin–DNase I complex. Nature 347, 37–44 ( 1990).
Whittaker, S. B-M., Boetzel, R., MacDonald, C., Lian, L-Y., Pommer, A.J., Reilly, A., James, R., Kleanthous, C. & Moore, G.R. NMR detection of slow conformational dynamics in an endonuclease toxin. J. Biol. NMR 12, 145–159 (1998).
Russo, A.A., Tong, L., Lee, J-O., Jeffrey, P.D. & Pavletich, N.P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16 (INK4a). Nature 395, 237–243 (1998).
Dalgaard, J.Z., Klar, A.J., Moser, M.J., Holley, W.R., Chatterjee, A. & Mian, I.S. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 25, 4626–4638 (1997).
Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A. & Nagata, S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43– 50 (1998).
Santoro, D.W. & Bolen, M.M. Unfolding free energy changes determined by the linear extrapolation method: unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry 27, 8063–8068 (1988).
Johnson, C.M. & Fersht, A. R. Protein stability as a function of denaturant concentration -the thermal-stability of barnase in the presence of urea. Biochemistry 34, 6795– 6804 (1995).
Otwinowski, Z. & Minor, W. Processing X-ray diffraction data collected in oscillation mode. Meth. Enz. 276, 307–326 (1996).
Sheldrick, G.M. Phase annealing in SHELX-90: direct methods for large structures. Acta Crystallogr. A 46, 467–473 (1990).
De la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Meth. Enz. 276, 472–494 (1997).
Abrahams, J.P. & Leslie, A.G.W. Methods used in the structural determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30– 42 (1996).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T., Kuriyan, J. & Karplus, M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Brünger, A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472– 475 (1992).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240– 255 (1997).
Navaza, J. AMoRe: an automated molecular replacement program package. Acta Crystallogr. A 50, 157–163 (1994).
Read, R.J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140 –149 (1986).
Laskowski, R.A., MacArthur, M.W., Moss, D. & Thornton, J.M. PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283– 292 (1993).
Acknowledgements
We thank the members of the UEA Colicin Research Group for valuable discussions, C. Garinot-Schneider (UEA) for early work on gel-shift experiments and A. Capaldi (Leeds) for help and advice regarding the folding experiments. We also thank the reviewers of this paper for their helpful comments. The work was generously supported by The Wellcome Trust and by the Biotechnology & Biological Sciences Research Council. N.F. was funded by the Medical Research Council.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kleanthous, C., Kühlmann, U., Pommer, A. et al. Structural and mechanistic basis of immunity toward endonuclease colicins . Nat Struct Mol Biol 6, 243–252 (1999). https://doi.org/10.1038/6683
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/6683
This article is cited by
-
Modeling endonuclease colicin-like bacteriocin operons as ‘genetic arms’ in plasmid-genome conflicts
Molecular Genetics and Genomics (2022)
-
Structural and functional insights into colicin: a new paradigm in drug discovery
Archives of Microbiology (2022)
-
Toxin import through the antibiotic efflux channel TolC
Nature Communications (2021)
-
Structural design principles for specific ultra-high affinity interactions between colicins/pyocins and immunity proteins
Scientific Reports (2021)
-
Suppressing the CRISPR/Cas adaptive immune system in bacterial infections
European Journal of Clinical Microbiology & Infectious Diseases (2017)