Abstract
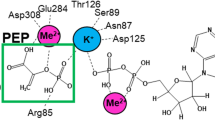
The crystal structure of L–lactate dehydrogenase from Bifidobacterium longum, determined to 2.5 Å resolution, contains a regular 1:1 complex of T– and R–state tetramers. A comparison of these two structures within the same crystal lattice and kinetical characterization of the T–R transition in solution provide an explanation for the molecular mechanism of allosteric activation. Substrate affinity is controlled by helix sliding between subunits which is triggered by the binding of the activator, fructose 1,6–bisphosphate. The proposed mechanism can explain activation by chemical modification and mutagenesis, as well as suggesting why vertebrate counterparts are not allosteric.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Perutz, M.F. Mechanism of Cooperativity and Allosteric Regulation in Proteins (Cambridge University Press, Cambridge, England, 1989).
Holbrook, J.J., Liljas, A., Steindel, S.J. & Rossmann, M.G. in The Enzymes (ed. Boyer, P.D.) 11, 191–292, (Academic, New York, 1975).
Garvie, E.I. Bacterial lactate dehydrogenases. Microbiol. Rev. 44, 106–139 (1980).
Minowa, T., Iwata, S., Sakai, H., Masaki, H. & Ohta, T. Bifidobacterium longum gene encoding L-lactate dehydrogenase and the primary structure of the enzyme: a new feature of the allosteric site. Gene 85, 161–168(1989).
Iwata, S., Minowa, T., Sakai H. & Ohta, T. Amino acid residues in the allosteric site of L-lactate dehydrogenase from Bifidobacterium longum. Agric. Biol. Chem. 53, 3365–3366 (1989).
de Vries, W., Gerbrandy, S.J. and Stouthamer, A.H. Carbohydrate metabolism in Bifidobacterium bifidum. Biochim. biophys. Acta 136, 415–425 (1967).
Ohta,T., Minowa, T., Yokota, K. & Iwata, S. Mechanism of allosteric transition of bacterial L-lactate dehydrogenase. Discuss. Faraday Soc. 93, 153–161 (1992).
Iwata, S., Minowa, T., Mikami, B., Morita, Y. & Ohta, T. Crystallization of and preliminary crystallographic data for allosteric L-lactate dehydrogenase fromBifidobacterium longum. J. Biochem. 106, 558–559 (1989).
Iwata, S. & Ohta, T. Molecular basis of allosteric activation of bacterial L-lactate dehydrogenase. J. molec. Biol. 230, 21–27 (1993).
Piontek, K., Chakrabarti, P., Schär, H.-P., Rossmann, M.G. & Zuber, H. Structure determination and refinement of Bacillus stearothermophiluslactate dehydrogenase. Proteins 7, 74–92 (1990).
Rossmann, M.G. et. al., Molecular symmetry axes and subunit interfaces in certain dehydrogenases. J. molec. Biol. 76, 533–537 (1973).
Clarke, A.R. et. al. Site-directed mutagenesis reveals role of mobile arginine residue in lactate dehydrogenase catalysis. Nature 324, 699–702 (1986).
Monod, J., Wyman, J. & Changeux, J.P. On the nature of allosteric transitions: A plausible model. J. molec. Biol. 12, 88–118 (1965).
Koide, S., Yokoyama, S., Matsuzawa, H., Miyazawa, T. & Ohta, T. Conformational equilibrium of an enzyme catalytic site in the allosteric transition. Biochemistry 31, 5362–5368 (1992).
Adams, M.J. et al. Structure of lactate dehydrogenase at 2.8 Å resolution. Nature 227, 1098–1103 (1970).
White, J.L. et al. A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J. molec. Biol. 102, 759–779 (1976).
Taguchi, H., Matsuzawa, H. & Ohta, T. L-lactate dehydrogenase from Thermus caldophilus GK24, an extremely thermophilic bacterium. Eur. J. Biochem. 145, 283–290 (1984).
Machida, M., Matsuzawa, H. & Ohta, T. Fructose 1,6-bisphosphate-dependent L-lactate dehydrogenase from Thermus aquaticus YT-1, an extreme thermophile: activation by citrate and modification reagents and comparison with Thermus caldophilus GK24 L-lactate dehydrogenase. J. Biochem. 97, 899–909 (1985).
Matsuzawa, H., Machida, M., Kunai, K., Ito, Y. & Ohta, T. Identification of an allosteric site residue of a fructose 1,6-bisphosphate-dependent L-lactate dehydrogenase of Thermus caldophilus GK24: production of non-allosteric form by protein engineering. FEBS Lett. 233, 375–387 (1988).
Schröder, G., Matsuzawa, H. & Ohta, T. Involvement of the conserved histidine-188 residue in the L-lactate dehydrogenase from Thermus caldophilus GK24 in allosteric regulation by fructose 1,6-bisphosphate. Biochim. biophys. Acta 152, 1236–1241 (1988).
Minowa, T. PhD thesis, A study on the activation mechanism of allosteric L-lactate dehydrogenase with protein engineering. (The University of Tokyo, Tokyo, Japan, 1990).
Eventoff, W. et al. Structural adaptations of lactate dehydrogenase isozymes. Proc. natn. Acad. Sci. U.S.A. 74, 2677–2681 (1977).
Hogrefe, H.H., Griffith, J.P., Rossmann, M.G. & Goldberg, E. Characterization of the antigenic sites on the refined 3 Å resolution structure of mouse testicular lactate dehydrogenase C4 . J. biol. Chem. 262, 13155–13162 (1987).
Grau, U.M., Trommer, W.E. & Rossmann, M.G. Structure of the active ternary complex of pig heart lactate dehydrogenase with S-lac-NAD+ at 2.7 Å resolution. J. molec. Biol. 151, 289–307 (1981).
Abad-zapatero, C., Griffith, J.P., Sussman, J.L. & Rossmann, M.G. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J. molec. Biol. 198, 445–467 (1987).
Wigley, D.B. et al. Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 Å resolution. J. molec. Biol. 223, 317–335 (1992).
Clarke, A.R., Atkinson, T., Campbell, J.W. & Holbrook, J.J. The assembly mechanism of the lactate dehydrogenase tetramer from Bacillus stearothermophilus: the equilibrium relationships between quaternary structure and the binding of fructose 1,6-bisphosphate. Biochim. biophys. Acta 829, 287–396 (1985).
Clarke, A.R. et al. A single amino acid substitution deregulates a bacterial lactate dehydrogenase and stabilizes its tetrameric structure. Biochim. biophys. Acta 913, 72–80 (1987).
Barford, D. & Johnson, L.N. The allosteric transition of glycogen phosphorylase. Nature 340, 609–616 (1989).
Schirmer, T. & Evans, P.R. Structural basis of the allosteric behaviour of phosphofructokinase. Nature 343, 140–145 (1990).
Perutz, M.F. Stereochemistry of cooperative effects in haemoglobin. Nature 228, 726–739 (1970).
Sprang, S.R. et al. Structural changes in glycogen phosphorylase induced by phosphorylation. Nature 336, 215–221 (1988).
Sakabe, N. A focusing Weissenberg camera with multi-layer-line screens for macromolecular crystallography.J. appl. Crystallogr. 16, 542–547 (1983).
Higashi, T. The processing of diffraction data taken on a screenless Weissenberg camera for macromolecular crystallography. J. appl. Crystallogr. 22, 9–18 (1989).
Bernstein, F.C. et al. The protein data bank: a computer based archival file for macromolecular structures. J. molec. Biol. 112, 535–542 (1977).
Abola, E.E., Bernstein, F.C., Bryant, S.H., Koetzle, T.F. & Weng, J. in Crystallographic databases — information content, software systems, scientific applications (ed. Alien, F. H. et al.) 107–132 (Data Commission of the International Union of Crystallography, Bonn/Cambridge/Chester, 1987).
Matsuura, Y. AUTOMR: an automatic processing program system for the molecular replacement method. J. appl. Crystallogr. 24, 1063–1066 (1991).
Brünger, A.T., Kuriyan, J. & Karplus, M., Crystallographic R-factor refinement by molecular dynamics. Science 235, 458–460 (1987)
Ramachandran, G.N. & Sasisekharan, V. Conformation of poly peptides and proteins. Adv. Prot. Chem. 23, 283–438 (1968)
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemistry of protein structures. J. appl. Crystallogr. 26, 283–291 (1993).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaad, M. Improved methods for building models in electron density maps and the location of errors in these models. Acta. Crystallogr. A47, 110–119 (1991).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. appl. Crystallogr. 24, 946–950 (1991).
Carson, M. RIBBONS 2.0. J. appl. Crystallogr. 24, 958–961 (1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Iwata, S., Kamata, K., Yoshida, S. et al. T and R states in the crystals of bacterial L–lactate dehydrogenase reveal the mechanism for allosteric control. Nat Struct Mol Biol 1, 176–185 (1994). https://doi.org/10.1038/nsb0394-176
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0394-176
This article is cited by
-
Identification of the first highly selective inhibitor of human lactate dehydrogenase B
Scientific Reports (2021)
-
Validation and quality assessment of macromolecular structures using complex network analysis
Scientific Reports (2019)
-
Thermal activation of ‘allosteric-like’ large-scale motions in a eukaryotic Lactate Dehydrogenase
Scientific Reports (2017)
-
Diverse allosteric and catalytic functions of tetrameric d-lactate dehydrogenases from three Gram-negative bacteria
AMB Express (2014)
-
50 years of allosteric interactions: the twists and turns of the models
Nature Reviews Molecular Cell Biology (2013)