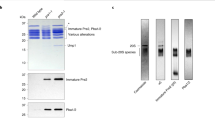
Abstract
HslVU is a new two-component protease in Escherichia coli composed of the proteasome-related peptidase HslV and the ATPase HslU. We have used electron microscopy and image analysis to examine the structural organization of HslV and HslU homo-oligomers and the active HslVU enzyme. Electron micrographs of HslV reveal ring-shaped particles, and averaging of top views reveal six-fold rotational symmetry, in contrast to other β-type proteasome subunits, which form rings with seven-fold symmetry. Side views of HslV show two rings stacked together; thus, HslV behaves as dodecamer. The ATPase HslU forms ring-shaped particles in the presence of ATP, AMP-PNP or ADP, suggesting that nucleotide binding, but not hydrolysis, is required for oligomerization. Subunit crosslinking, STEM mass estimation, and analysis of HslU top views indicate that HsIU exists both as hexameric and heptameric rings. With AMP-PNP present, maximal proteolytic activity is observed with a molar ratio of HsIU to HsIV subunits of 1:1, and negative staining electron microscopy shows that HsIV and HsIU form cylindrical four-ring structures in which the HslV dodecamer is flanked at each end by a HslU ring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Parsell, D.A. & Lindquist, S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Ann. Rev. Genet. 27, 437–496 (1993).
Goldberg, A.L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur. J. Biochem 203, 9–923 (1992).
Hayes, S.A. & Dice, J.F. Roles of molecular chaperones in protein degradation. J. Cell Biol. 132, 255–258 (1996).
Goff, S.A., Casson, L.P. & Goldberg, A.L. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the Ion gene in Escherichia coli. Proc.Natl.Acad.Sci. USA 81, 6647–6651 (1984).
Kroh, H.E. & Simon, L.D. CIpP component of Clp protease is the sigma 32-dependent heat shock protein F21.5. J. Bacteriol. 172, 6026–6034 (1990).
Wojtkowiak, D., Georgopoulos, C. & Zylicz, M. Isolation and characterization of CIpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J. Biol. Chem. 268, 22609–22617 (1993).
Tomoyasu, T. et al. Escherichia coll FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14, 2551–2560 (1995).
Rohrwild, M. et al. HsIV-HsIU- a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA 93, 5808–5813 (1996).
Yoo, S.J. et al Purification and characterization of the heat shock proteins HsIV and HsIU that form a new ATP-dependent protease in Escherichia coli. J. Biol.Chem. 271, 14035–14040 (1996).
Fleischmann, R.D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Highlander, S.K., Wickersham, E.A., Garza, O. & Weinstock, G.M. Expression of the Pasteurella haemolytica leukotoxin is inhibited by a locus that encodes an ATP-binding cassette homolog. Infect. Immun. 61, 3942–3951 (1993).
Lupas, A., Zwickl, P. & Baumeister, W. Proteasome sequences in eubacteria. Trends Biochem. Sci. 19, 533–534 (1994).
Slack, F.J., Serror, P., Joyce, E. & Sonenshein, A.L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15, 689–702 (1995).
Becker, J. & Brendel, M. Molecular characterization of the xerC gene of Lactobacillus Leichmannii encoding a site-specific recombinase and two adjacent heat shock genes. Curr. Microbiol. 32, 232–236 (1996).
Schirmer, E.G., Glover, J.R., Singer, M.A. & Lindquist, S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21, 289–296 (1996).
Chuang, S.E., Burland, V., Plunkett, G.d., Daniels, D.L. & Blattner, F.R. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coll. Gene 134, 1–6 (1993).
Maurizi, M.R., Thompson, M.W., Singh, S.K. & Kirn, S.H., Clp protease from Escherichia coli. Meths. Enzymol. 244, 314–331 (1994).
Dahlmann, B. et al. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Letters 251, 125–131 (1989).
Tamura, T. et al. The first characterization of a eubacterial proteasome - the 20S complex of Rhodococcus. Curr. Biol. 5, 766–774 (1995).
Tanahashi, N., Tsurumi, C., Tamura, T. & Tanaka, K. Molecular structures of 20S and 26S proteasomes. Enzyme Prot. 47, 241–251 (1993).
Lupas, A., Koster, A.J. & Baumeister, W. Structural features of 26S and 20S proteasomes. Enzyme Prot. 47, 252–273 (1993).
Pühler, G. et al. Subunit stoichiometry and three-dimensional arrangement in proteasomes from Thermoplasma acidophilum. EMBO J. 11, 1607–1616 (1992).
Waxman, L, Fagan, J.M. & Goldberg, A.L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. Biol. Chem. 262, 2451–2457 (1987).
Hough, R., Pratt, G. & Rechsteiner, M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J. Biol. Chem. 262, 8303–8313 (1987).
Ganoth, D., Leshinsky, E., Eytan, E. & Hershko, A. A multicomponent system that degrades proteins conjugated to ubiquitin: resolution of factors and evidence for ATP-dependent complex formation. J. Biol. Chem. 263, 12412–12419 (1988).
Hershko, A. & Ciechanover, A. The ubiquitin system for protein degradation. Ann. Rev. Biochem. 61, 761–807 (1992).
Peters, J.M., Cejka, Z., Harris, J.R., Kleinschmidt, J.A. & Baumeister, W. Structural features of the 26-S proteasome complex. J. Mol. Biol. 234, 932–937 (1993).
Coux, O., Tanaka, K. & Goldberg, A.L. Structure and functions of the 20S and 26S proteasomes. Ann. Rev. Biochem 65, 801–647 (1996).
Seemüller, E., Lupas, A. & Baumeister, W. Autocatalytic processing of the 20S proteasome. Nature 382, 468–470 (1996).
Kessel, M. et al. Homology in structural organization between E. coli CIpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol. 250, 587–594 (1995).
Löwe, J. et al. Crystal structure of the 20S proteasome from the archaeon I acidophilum at 3.4 A resolution. Science 268, 533–539 (1995).
Müller, S., Goldie, K.N., Bürki, R., Häring, R. & Engel, A. Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy 46, 317 (1992).
Engel, A. & Colliex, C. Application of scanning transmission electron microscopy to the study of biological structure. Curr. Opin. Biotechnol. 4, 403–411 (1993).
Gottesman, S. et al. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. USA 87, 3513–3517 (1990).
Zwickl, P., Kleinz, J. & Baumeister, W. Critical elements in proteasome assembly. Nature Struct. Biol. 1, 765–770 (1994).
Nederlof, P.M., Wang, H.R. & Baumeister, W. Nuclear localization signals of human and Thermoplasma cc subunits are functional in vitro. Proc. Natl. Acad. Sci. USA 92, 12060–12064 (1995).
Shin, D.H., Lee, C.S., Chung, C.H. & Suh, S.W. Molecular symmetry of the CIpP component of the ATP-dependent Clp Protease, an Escherichia coli homolog of the 20S proteasome. J. Mol. Biol. 262, 71–76 (1996).
Weissman, J.S., Sigler, P.B. & Horwich, A.L. From the cradle to the grave: ring complexes in the life of a protein. Science 268, 523–524 (1995).
Kelman, Z., Finkelstein, J. & O′Donnell, M. Protein structure. Why have six-fold symmetry? Curr. Biol. 5, 1239–1242 (1995).
Wawrzynow, A. et al. The CIpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the CIpP-CIpX protease, is a novel molecular chaperone. EMBOJ. 14, 1867–1877 (1995).
Wickner, S. et al. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl.Acad.Sci. USA 91, 12218–12222 (1994).
Missiakas, D., Schwager, F., Betton, J.-M., Georgopoulos, C. & Raina, S. Identification and characterization of HslV HslU (CIpQ CIpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J., in press.
Hegerl, R. & Altbauer, A. The “EM” program system. Ultramicroscopy 9, 109–116 (1982).
Saxton, W.O., Pitt, T.J. & Horner, M. Digital image processing: the Semper system. Ultramicroscopy 4, 343–354 (1979).
Frank, J., Bretaudiere, J.P., Carazo, J.M., Verschoor, A. & Wagenknecht, T. Classification of images of biomolecular assemblies: a study of ribosomes and ribosomal subunits of Escherichia coli. J. Microscopy 150, 99–115 (1988).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rohrwild, M., Pfeifer, G., Santarius, U. et al. The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat Struct Mol Biol 4, 133–139 (1997). https://doi.org/10.1038/nsb0297-133
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0297-133
This article is cited by
-
Protein degradation control and regulation of bacterial survival and pathogenicity: the role of protein degradation systems in bacteria
Molecular Biology Reports (2021)
-
Assessing heterogeneity in oligomeric AAA+ machines
Cellular and Molecular Life Sciences (2017)
-
Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes
BMC Genomics (2010)
-
Plant Hsp100/ClpB-like proteins: poorly-analyzed cousins of yeast ClpB machine
Plant Molecular Biology (2010)
-
Proteasome-Related HslU and HslV Genes Typical of Eubacteria Are Widespread in Eukaryotes
Journal of Molecular Evolution (2006)