Abstract
Renal cell carcinoma (RCC) is a constellation of malignancies of different histological subtypes arising from the renal parenchyma. The clear cell histological subtype (ccRCC) accounts for around 75% of RCCs and is characterized by distinct genetic abnormalities, of which the loss of function of the von Hippel–Lindau (VHL) tumor suppressor gene is the most common. Inactivation of other tumor suppressor genes such as SETD2, KDM6A, KDM5C and PBRM1 has been reported in ccRCC—notably, the proteins encoded by these genes are involved in histone and chromatin regulation. Furthermore, the PBRM1 and SETD2 genes are located on the short arm of chromosome 3 near the VHL locus. Chromatin and histones modify gene expression and, as a consequence, their function is tightly regulated. Data from RNA interference (RNAi) assays suggest that loss of function of PBRM1 drives proliferation and growth of ccRCC, but the clinical relevance of this is unclear and restoring the function of these genes for therapeutic purposes is likely to be challenging. An improved understanding of histone and chromatin regulation in RCC biology and the consequences of intratumor heterogeneity might identify novel targets in RCC and present alternative therapeutic opportunities.
Key Points
-
Clear cell renal cell carcinoma (ccRCC) accounts for about 75% of all RCCs
-
Newly reported mutations of tumor suppressor genes SETD2, KDM6A and KDM5C, which encode proteins involved in histone modification, have been shown to occur in ccRCC
-
Chromosomal locus 3p21, where VHL, BAP1, SETD2 and PBRM1 reside, highlights a hot spot for mutations of genes involved in ccRCC
-
Newly identified somatic mutations in histone modifying enzymes suggest that chromatin and histone modifications might have important roles in the pathogenesis of ccRCC
-
A better understanding of the roles of histone modification provides a potential therapeutic target in ccRCC
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jemal, A. et al. Global cancer statistics, 2010. CA Cancer J. Clin. 61, 69–90 (2011).
Mathew, A., Devesa, S. S., Fraumeni, J. F. Jr & Chow, W. H. Global increases in kidney cancer incidence, 1973–1992. Eur. J. Cancer Prev. 11, 171–178 (2002).
Sun, M. et al. Age-adjusted incidence, mortality, and survival rates of stage-specific renal cell carcinoma in North America: a trend analysis. Eur. Urol. 59, 135–141 (2011).
Lam, J. S., Leppert, J. T., Belldegrun, A. S. & Figlin, R. A. Novel approaches in the therapy of metastatic renal cell carcinoma. World J. Urol. 23, 202–212 (2005).
Fisher R. A. et al. Observation prior to systemic therapy in patients with metastatic renal cell carcinoma in the kinase inhibitor era. J. Clin. Oncol. 29, e241–e242 (2011).
Lopez-Beltran, A., Scarpelli, M., Montironi, R. & Kirkali, Z. 2004 WHO classification of the renal tumors of the adults. Eur. Urol. 49, 798–805 (2006).
Storkel, S. et al. Classification of renal cell carcinoma: Workgroup No. 1. Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC). Cancer 80, 987–989 (1997).
Renshaw, A. A. Subclassification of renal cell neoplasms: an update for the practising pathologist. Histopathology 41, 283–300 (2002).
Kim, W. Y. & Kaelin, W. G. Role of VHL gene mutation in human cancer. J. Clin. Oncol. 22, 4991–5004 (2004).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Schmidt, L. et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 16, 68–73 (1997).
Alam, N. A. et al. Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum. Mol. Genet. 12, 1241–1252 (2003).
Alam, N. A. et al. Missense mutations in fumarate hydratase in multiple cutaneous and uterine leiomyomatosis and renal cell cancer. J. Mol. Diagn. 7, 437–443 (2005).
Tomlinson, I. P. et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410 (2002).
Hong, S. B. et al. Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-beta signaling. Mol. Cancer 9, 160 (2010).
Toro, J. R. Birt-Hogg-Dubé syndrome. In GeneReviews (eds Pagon, R. A., Bird, T. D., Dolan, C. R. & Stephens, K.) (2008).
Schmidt, L. S. et al. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dubé syndrome. Am. J. Hum. Genet. 76, 1023–1033 (2005).
Khoo, S. K. et al. Inactivation of BHD in sporadic renal tumors. Cancer Res. 63, 4583–4587 (2003).
Argani, P. et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am. J. Surg. Pathol. 31, 1149–1160 (2007).
Ross, H. & Argani, P. Xp11 translocation renal cell carcinoma. Pathology 42, 369–373 (2010).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41, 521–523 (2009).
Hurst, F. P. et al. Incidence, predictors and associated outcomes of renal cell carcinoma in long-term dialysis patients. Urology 77, 1271–1276 (2011).
Strumberg, D. Efficacy of sunitinib and sorafenib in non-clear cell renal cell carcinoma: results from expanded access studies. J. Clin. Oncol. 26, 3469–3471 (2008).
Escudier, B. et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N. Engl. J. Med. 356, 125–134, (2007).
Sternberg, C. N. et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J. Clin. Oncol. 28, 1061–1068 (2010).
Coppin, C. et al. Immunotherapy for advanced renal cell cancer. Cochrane Database of Systematic Reviews, Issue 1. Art No.: CD001425. http://dx.doi.org/10.1002/14651858.CD001425.pub2 (2005).
Yagoda, A., Petrylak, D. & Thompson, S. Cytotoxic chemotherapy for advanced renal cell carcinoma. Urol. Clin. North Am. 20, 303–321 (1993).
Rini, B. I. Metastatic renal cell carcinoma: many treatment options, one patient. J. Clin. Oncol. 27, 3225–3234 (2009).
Horiuchi, A. et al. Hypoxia-induced changes in the expression of VEGF, HIF-1 alpha and cell cycle-related molecules in ovarian cancer cells. Anticancer Res. 22, 2697–2702 (2002).
Harada, H. et al. The Akt/mTOR pathway assures the synthesis of HIF-1alpha protein in a glucose- and reoxygenation-dependent manner in irradiated tumors. J. Biol. Chem. 284, 5332–5342 (2009).
Rini, B. I., Campbell, S. C. & Escudier, B. Renal cell carcinoma. Lancet 373, 1119–1132 (2009).
Motzer, R. J. et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 27, 3584–3590 (2009).
Motzer, R. J. et al. Phase 3 trial of everolimus for metastatic renal cell carcinoma: final results and analysis of prognostic factors. Cancer 116, 4256–4265 (2010).
Swanton, C. et al. Predictive biomarker discovery through the parallel integration of clinical trial and functional genomics datasets. Genome Med. 2, 53 (2010).
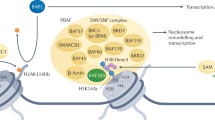
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Muhlbauer, B. et al. Magnesium-L-aspartate-HCl and magnesium-oxide: bioavailability in healthy volunteers. Eur. J. Clin. Pharmacol. 40, 437–438 (1991).
Geiman, T. M. & Robertson, K. D. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J. Cell Biochem. 87, 117–125 (2002).
Doerfler, W. et al. Eukaryotic DNA methylation: facts and problems. FEBS Lett. 268, 329–333 (1990).
Nojima, D. et al. CpG methylation of promoter region inactivates E-cadherin gene in renal cell carcinoma. Mol. Carcinog. 32, 19–27 (2001).
Morris, M. R. et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 29, 2104–2117 (2010).
Cooper, G. M. The Cell (Boston University, Sunderland: Sinauer Associates, 2000).
Paranjape, S. M., Kamakaka, R. T. & Kadonaga, J. T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63, 265–297 (1994).
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997).
Horn, P. J. & Peterson, C. L. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science 297, 1824–1827 (2002).
Kurdistani, S. K. Histone modifications in cancer biology and prognosis. Prog. Drug Res. 67, 91–106 (2011).
Sims, R. J. 3rd, Nishioka, K. & Reinberg, D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19, 629–639 (2003).
Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606 (1998).
Bartova, E., Krejci, J., Harnicarova, A., Galiova, G. & Kozubek, S. Histone modifications and nuclear architecture: a review. J. Histochem. Cytochem. 56, 711–721 (2008).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Bird, A. Perceptions of epigenetics. Nature 447, 396–398 (2007).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Bannister, A. J., Schneider, R. & Kouzarides, T. Histone methylation: dynamic or static? Cell 109, 801–806 (2002).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Di Lorenzo, A. & Bedford, M. T. Histone arginine methylation. FEBS Lett. 585, 2024–2031 (2011).
Lee, D. Y., Teyssier, C., Strahl, B. D. & Stallcup, M. R. Role of protein methylation in regulation of transcription. Endocr. Rev. 26, 147–170 (2005).
Berger, S. L. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007).
Ruthenburg, A. J., Allis, C. D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007).
Klose, R. J. & Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 (2007).
Hess, J. L. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol. Med. 10, 500–507 (2004).
Barrett, A. et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int. J. Cancer 121, 265–275 (2007).
Xiang, Y. et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl Acad. Sci. USA 104, 19226–19231 (2007).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003).
Simon, J. A. & Lange, C. A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647, 21–29 (2008).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Wagener, N. et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int. J. Cancer 123, 1545–1550 (2008).
Ougolkov, A. V., Bilim, V. N. & Billadeau, D. D. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin. Cancer Res. 14, 6790–6796 (2008).
Chase, A. & Cross, N. C. Aberrations of EZH2 in cancer. Clin. Cancer Res. 17, 2613–2618 (2011).
Beyer, S., Kristensen, M. M., Jensen, K. S., Johansen, J. V. & Staller, P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283, 36542–36552 (2008).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Martinez-Garcia, E. & Licht, J. D. Deregulation of H3K27 methylation in cancer. Nat. Genet. 42, 100–101 (2010).
Bannister, A. J. et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280, 17732–17736 (2005).
Huang, J. et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 285, 9636–9641 (2010).
Yamane, K. et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25, 801–812 (2007).
Reisman, D., Glaros, S. & Thompson, E. A. The SWI/SNF complex and cancer. Oncogene 28, 1653–1668 (2009).
Xia, W. et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 68, 1667–1674 (2008).
Burrows, A. E., Smogorzewska, A. & Elledge, S. J. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl Acad. Sci. USA 107, 14280–14285 (2010).
Janssen, A., Kops, G. J. & Medema, R. H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. USA 106, 19108–19113 (2009).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Kenneth, N. S., Mudie, S., van Uden, P. & Rocha, S. SWI/SNF regulates the cellular response to hypoxia. J. Biol. Chem. 284, 4123–4131 (2009).
Guo, G. et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 44, 17–19 (2012).
Morita, R. et al. Common regions of deletion on chromosomes 5q, 6q, and 10q in renal cell carcinoma. Cancer Res. 51, 5817–5820 (1991).
Flaig, T. W. et al. Safety and efficacy of the combination of erlotinib and sirolimus for the treatment of metastatic renal cell carcinoma after failure of sunitinib or sorafenib. Br. J. Cancer 103, 796–801 (2010).
Bukowski, R. M. et al. Randomized phase II study of erlotinib combined with bevacizumab compared with bevacizumab alone in metastatic renal cell cancer. J. Clin. Oncol. 25, 4536–4541 (2007).
Jermann, M. et al. A phase II, open-label study of gefitinib (IRESSA) in patients with locally advanced, metastatic, or relapsed renal-cell carcinoma. Cancer Chemother. Pharmacol. 57, 533–539 (2006).
Ravaud, A. et al. Lapatinib versus hormone therapy in patients with advanced renal cell carcinoma: a randomized phase III clinical trial. J. Clin. Oncol. 26, 2285–2291 (2008).
Shand, N. et al. A phase 1–2 clinical trial of gene therapy for recurrent glioblastoma multiforme by tumor transduction with the herpes simplex thymidine kinase gene followed by ganciclovir. GLI328 European-Canadian Study Group. Hum. Gene Ther. 10, 2325–2335 (1999).
Buchner, A. et al. Phase 1 trial of allogeneic gene-modified tumor cell vaccine RCC-26/CD80/IL-2 in patients with metastatic renal cell carcinoma. Hum. Gene Ther. 21, 285–297 (2010).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Roylance, R. et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomarkers Prev. 20, 2183–2194 (2011).
Moch, H. et al. Intratumoral heterogeneity of von Hippel-Lindau gene deletions in renal cell carcinoma detected by fluorescence in situ hybridization. Cancer Res. 58, 2304–2309 (1998).
Li, G. et al. Different DNA ploidy patterns for the differentiation of common subtypes of renal tumors. Cell Oncol. 27, 51–56 (2005).
Ljungberg, B., Mehle, C., Stenling, R. & Roos, G. Heterogeneity in renal cell carcinoma and its impact no prognosis--a flow cytometric study. Br. J. Cancer 74, 123–127 (1996).
Zama, I. N. et al. Sunitinib rechallenge in metastatic renal cell carcinoma patients. Cancer 116, 5400–5406 (2010).
Schifitto, G. et al. Valproic acid adjunctive therapy for HIV-associated cognitive impairment: a first report. Neurology 66, 919–921 (2006).
Kramer, O. H., Knauer, S. K., Zimmermann, D., Stauber, R. H. & Heinzel, T. Histone deacetylase inhibitors and hydroxyurea modulate the cell cycle and cooperatively induce apoptosis. Oncogene 27, 732–740 (2008).
Swoboda, K. J. et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS ONE 4, e5268, (2009).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Grant, C. et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev. Anticancer Ther. 10, 997–1008 (2010).
Olsen, E. A. et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 25, 3109–3115 (2007).
Vansteenkiste, J. et al. Early phase II trial of oral vorinostat in relapsed or refractory breast, colorectal, or non-small cell lung cancer. Invest. New Drugs 26, 483–488 (2008).
Modesitt, S. C., Sill, M., Hoffman, J. S. & Bender, D. P. A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol. Oncol. 109, 182–186 (2008).
Klimek, V. M. et al. Tolerability, pharmacodynamics, and pharmacokinetics studies of depsipeptide (romidepsin) in patients with acute myelogenous leukemia or advanced myelodysplastic syndromes. Clin. Cancer Res. 14, 826–832 (2008).
Schrump, D. S. et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin. Cancer Res. 14, 188–198 (2008).
Nagji, A. S., Cho, S. H., Liu, Y., Lee, J. K. & Jones, D. R. Multigene expression-based predictors for sensitivity to Vorinostat and Velcade in non-small cell lung cancer. Mol. Cancer Ther. 9, 2834–2843 (2010).
Chen, C. S., Weng, S. C., Tseng, P. H. & Lin, H. P. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J. Biol. Chem. 280, 38879–38887 (2005).
Traynor, A. M. et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J. Thorac. Oncol. 4, 522–526 (2009).
Garber, K. HDAC inhibitors overcome first hurdle. Nat. Biotechnol. 25, 17–19 (2007).
Tan, J. et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21, 1050–1063 (2007).
Miranda, T. B. et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8, 1579–1588 (2009).
Zhou, J. et al. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood 118, 2830–2839 (2011).
Cha, T. L. et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 (2005).
Shi, L. et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl Acad. Sci. USA 108, 7541–7546 (2011).
Duns, G. et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 70, 4287–4291 (2010).
Author information
Authors and Affiliations
Contributions
J. Larkin, X. Y. Goh and M. Vetter contributed equally to the writing of this manuscript, researching data for the article, writing the manuscript and making a substantial contribution to discussion of content, as well as reviewing and editing the manuscript before submission. L. Pickering reviewed and edited the article before submission. C. Swanton researched data for the article, made a substantial contribution to discussion of content and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Larkin, J., Goh, X., Vetter, M. et al. Epigenetic regulation in RCC: opportunities for therapeutic intervention?. Nat Rev Urol 9, 147–155 (2012). https://doi.org/10.1038/nrurol.2011.236
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrurol.2011.236
This article is cited by
-
ADAMTS9-AS1 inhibits tumor growth and drug resistance in clear cell renal cell carcinoma via recruiting HuR to enhance ADAMTS9 mRNA stability
Cancer Nanotechnology (2023)
-
SNP-mediated lncRNA-ENTPD3-AS1 upregulation suppresses renal cell carcinoma via miR-155/HIF-1α signaling
Cell Death & Disease (2021)
-
The diverse roles of SPOP in prostate cancer and kidney cancer
Nature Reviews Urology (2020)
-
Simultaneous targeted activation of Notch1 and Vhl-disruption in the kidney proximal epithelial tubular cells in mice
Scientific Reports (2016)
-
Conditional inactivation of the mouse von Hippel–Lindau tumor suppressor gene results in wide-spread hyperplastic, inflammatory and fibrotic lesions in the kidney
Oncogene (2015)