Abstract
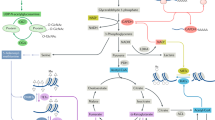
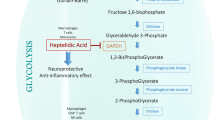
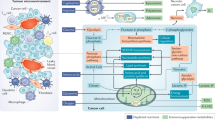
All immune cells depend on specific and efficient metabolic pathways to mount an appropriate response. Over the past decade, the field of immunometabolism has expanded our understanding of the various means by which cells modulate metabolism to achieve the effector functions necessary to fight infection or maintain homeostasis. Harnessing these metabolic pathways to manipulate inappropriate immune responses as a therapeutic strategy in cancer and autoimmunity has received increasing scrutiny by the scientific community. Fine tuning immunometabolism to provide the desired response, or prevent a deleterious response, is an attractive alternative to chemotherapy or overt immunosuppression. The various metabolic pathways used by immune cells in rheumatoid arthritis, systemic lupus erythematosus and osteoarthritis offer numerous opportunities for selective targeting of specific immune cell subsets to manipulate cellular metabolism for therapeutic benefit in these rheumatologic diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Yildirim-Toruner, C. & Diamond, B. Current and novel therapeutics in the treatment of systemic lupus erythematosus. J. Allergy Clin. Immunol. 127, 303–312 (2011).
Kahlenberg, J. M. & Fox, D. A. Advances in the medical treatment of rheumatoid arthritis. Hand Clin. 27, 11–20 (2011).
Sokolove, J. & Lepus, C. M. Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther. Adv. Musculoskelet. Dis. 5, 77–94 (2013).
Hirahara, K., Schwartz, D., Gadina, M., Kanno, Y. & O'Shea, J. J. Targeting cytokine signaling in autoimmunity: back to the future and beyond. Curr. Opin. Immunol. 43, 89–97 (2016).
Tanaka, Y., Nakayamada, S. & Okada, Y. Osteoblasts and osteoclasts in bone remodeling and inflammation. Curr. Drug Targets Inflamm. Allergy 4, 325–328 (2005).
Ikeda, K. & Takeshita, S. The role of osteoclast differentiation and function in skeletal homeostasis. J. Biochem. 159, 1–8 (2016).
O'Neill, L. A., Kishton, R. J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016).
Buck, M. D., O'Sullivan, D. & Pearce, E. L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
Knowles, H. J. Hypoxic regulation of osteoclast differentiation and bone resorption activity. Hypoxia (Auckl.) 3, 73–82 (2015).
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Sukumar, M. et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2016).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Kawalekar, O. U. et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016).
Moreau, H. D. & Bousso, P. Visualizing how T cells collect activation signals in vivo. Curr. Opin. Immunol. 26, 56–62 (2014).
Frauwirth, K. A. et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002).
Jacobs, S. R. et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Gerriets, V. A. et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015).
Parry, R. V. et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 25, 9543–9553 (2005).
Patsoukis, N. et al. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci. Signal. 5, ra46 (2012).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Perl, A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat. Rev. Rheumatol. 12, 169–182 (2016).
Caro-Maldonado, A. et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol. 192, 3626–3636 (2014).
Blair, D., Dufort, F. J. & Chiles, T. C. Protein kinase Cβ is critical for the metabolic switch to glycolysis following B-cell antigen receptor engagement. Biochem. J. 448, 165–169 (2012).
Cho, S. H. et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature 537, 234–238 (2016).
Iwata, T. N. et al. Conditional disruption of Raptor reveals an essential role for mTORC1 in B cell development, survival, and metabolism. J. Immunol. 197, 2250–2260 (2016).
Jellusova, J. & Rickert, R. C. The PI3K pathway in B cell metabolism. Crit. Rev. Biochem. Mol. Biol. 51, 359–378 (2016).
van der Windt, G. J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl Acad. Sci. USA 110, 14336–14341 (2013).
Gubser, P. M. et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14, 1064–1072 (2013).
Na, Y. R. et al. GM-CSF induces inflammatory macrophages by regulating glycolysis and lipid metabolism. J. Immunol. 197, 4101–4109 (2016).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 (2016).
Semba, H. et al. HIF-1α–PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat. Commun. 7, 11635 (2016).
O'Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Freemerman, A. J. et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 (2014).
Everts, B. et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15, 323–332 (2014).
Pearce, E. J. & Everts, B. Dendritic cell metabolism. Nat. Rev. Immunol. 15, 18–29 (2015).
Krawczyk, C. M. et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 (2010).
Cheng, S. C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Johnson, A. R. et al. Metabolic reprogramming through fatty acid transport protein 1 (FATP1) regulates macrophage inflammatory potential and adipose inflammation. Mol. Metab. 5, 506–526 (2016).
Burr, D. B. & Gallant, M. A. Bone remodelling in osteoarthritis. Nat. Rev. Rheumatol. 8, 665–673 (2012).
Indo, Y. et al. Metabolic regulation of osteoclast differentiation and function. J. Bone Miner. Res. 28, 2392–2399 (2013).
Morten, K. J., Badder, L. & Knowles, H. J. Differential regulation of HIF-mediated pathways increases mitochondrial metabolism and ATP production in hypoxic osteoclasts. J. Pathol. 229, 755–764 (2013).
Nasi, A. et al. Dendritic cell reprogramming by endogenously produced lactic acid. J. Immunol. 191, 3090–3099 (2013).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Shi, L. Z. et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Waickman, A. T. & Powell, J. D. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 249, 43–58 (2012).
Boothby, M. Signaling in T cells — is anything the m(a)TOR with the picture(s)? F1000Res. 5, 191 (2016).
McDonald, G. et al. Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J. Clin. Invest. 124, 712–724 (2014).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Gerriets, V. A. et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17, 1459–1466 (2016).
Basu, S., Hubbard, B. & Shevach, E. M. Foxp3-mediated inhibition of Akt inhibits Glut1 (glucose transporter 1) expression in human T regulatory cells. J. Leukoc. Biol. 97, 279–283 (2015).
Beier, U. H. et al. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 29, 2315–2326 (2015).
Macintyre, A. N. et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 (2014).
Nakaya, M. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 (2014).
Sinclair, L. V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Zeng, H. & Chi, H. Metabolic control of regulatory T cell development and function. Trends Immunol. 36, 3–12 (2015).
Procaccini, C. et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity 33, 929–941 (2010).
De Rosa, V. et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 16, 1174–1184 (2015).
Shrestha, S. et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat. Immunol. 16, 178–187 (2015).
Huynh, A. et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196 (2015).
Park, Y. et al. TSC1 regulates the balance between effector and regulatory T cells. J. Clin. Invest. 123, 5165–5178 (2013).
Ray, J. P. et al. The interleukin-2–mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity 43, 690–702 (2015).
Zeng, H. et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45, 540–554 (2016).
Covarrubias, A. J., Aksoylar, H. I. & Horng, T. Control of macrophage metabolism and activation by mTOR and Akt signaling. Semin. Immunol. 27, 286–296 (2015).
Weichhart, T., Hengstschlager, M. & Linke, M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 15, 599–614 (2015).
Bengsch, B. et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 (2016).
Yan, B. et al. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod. Rheumatol. 26, 914–922 (2016).
Guleria, A. et al. NMR based serum metabolomics reveals a distinctive signature in patients with lupus nephritis. Sci. Rep. 6, 35309 (2016).
Wu, T. et al. Metabolic disturbances associated with systemic lupus erythematosus. PLoS ONE 7, e37210 (2012).
Wahl, D. R. et al. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus 19, 1492–1501 (2010).
Nagy, G., Koncz, A. & Perl, A. T cell activation-induced mitochondrial hyperpolarization is mediated by Ca2+- and redox-dependent production of nitric oxide. J. Immunol. 171, 5188–5197 (2003).
Nagy, G., Koncz, A., Fernandez, D. & Perl, A. Nitric oxide, mitochondrial hyperpolarization, and T cell activation. Free Radic. Biol. Med. 42, 1625–1631 (2007).
Gergely, P. Jr et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 46, 175–190 (2002).
Morel, L., Blenman, K. R., Croker, B. P. & Wakeland, E. K. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl Acad. Sci. USA 98, 1787–1792 (2001).
Perry, D. J. et al. Murine lupus susceptibility locus Sle1c2 mediates CD4+ T cell activation and maps to estrogen-related receptor γ. J. Immunol. 189, 793–803 (2012).
Eichner, L. J. et al. miR-378* mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 12, 352–361 (2010).
Bensinger, S. J. et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134, 97–111 (2008).
Kidani, Y. & Bensinger, S. J. LXR and PPAR as integrators of lipid homeostasis and immunity. Immunol. Rev. 249, 72–83 (2012).
Tso, T. K., Huang, H. Y., Chang, C. K., Liao, Y. J. & Huang, W. N. Clinical evaluation of insulin resistance and β-cell function by the homeostasis model assessment in patients with systemic lupus erythematosus. Clin. Rheumatol. 23, 416–420 (2004).
Gabriel, C. L. et al. Autoimmune-mediated glucose intolerance in a mouse model of systemic lupus erythematosus. Am. J. Physiol. Endocrinol. Metab. 303, E1313–E1324 (2012).
Wilhelm, A. J. & Major, A. S. Accelerated atherosclerosis in SLE: mechanisms and prevention approaches. Int. J. Clin. Rheumtol. 7, 527–539 (2012).
Saucillo, D. C., Gerriets, V. A., Sheng, J., Rathmell, J. C. & Maciver, N. J. Leptin metabolically licenses T cells for activation to link nutrition and immunity. J. Immunol. 192, 136–144 (2014).
Gerriets, V. A. et al. Leptin directly promotes T-cell glycolytic metabolism to drive effector T-cell differentiation in a mouse model of autoimmunity. Eur. J. Immunol. 46, 1970–1983 (2016).
Lourenco, E. V., Liu, A., Matarese, G. & La Cava, A. Leptin promotes systemic lupus erythematosus by increasing autoantibody production and inhibiting immune regulation. Proc. Natl Acad. Sci. USA 113, 10637–10642 (2016).
Yang, Z., Fujii, H., Mohan, S. V., Goronzy, J. J. & Weyand, C. M. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J. Exp. Med. 210, 2119–2134 (2013).
Sena, L. A. et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013).
Yang, Z. et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci. Transl Med. 8, 331ra38 (2016).
Fearon, U., Canavan, M., Biniecka, M. & Veale, D. J. Hypoxia, mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat. Rev. Rheumatol. 12, 385–397 (2016).
Hong, Y. H. & Kong, E. J. (18F)Fluoro-deoxy-D-glucose uptake of knee joints in the aspect of age-related osteoarthritis: a case-control study. BMC Musculoskelet. Disord. 14, 141 (2013).
Courties, A., Gualillo, O., Berenbaum, F. & Sellam, J. Metabolic stress-induced joint inflammation and osteoarthritis. Osteoarthritis Cartilage 23, 1955–1965 (2015).
Franke, S. et al. Advanced glycation end products affect growth and function of osteoblasts. Clin. Exp. Rheumatol. 29, 650–660 (2011).
Martinez-Calatrava, M. J. et al. RANKL synthesized by articular chondrocytes contributes to juxta-articular bone loss in chronic arthritis. Arthritis Res. Ther. 14, R149 (2012).
Jin, Z., Wei, W., Yang, M., Du, Y. & Wan, Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 20, 483–498 (2014).
Lemma, S. et al. Energy metabolism in osteoclast formation and activity. Int. J. Biochem. Cell Biol. 79, 168–180 (2016).
Vander Heiden, M. G. Exploiting tumor metabolism: challenges for clinical translation. J. Clin. Invest. 123, 3648–3651 (2013).
O'Sullivan, D. & Pearce, E. L. Targeting T cell metabolism for therapy. Trends Immunol. 36, 71–80 (2015).
Wheaton, W. W. et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife 3, e02242 (2014).
Son, H. J. et al. Metformin attenuates experimental autoimmune arthritis through reciprocal regulation of Th17/Treg balance and osteoclastogenesis. Mediators Inflamm. 2014, 973986 (2014).
Bian, L. et al. Dichloroacetate alleviates development of collagen II-induced arthritis in female DBA/1 mice. Arthritis Res. Ther. 11, R132 (2009).
Ostroukhova, M. et al. The role of low-level lactate production in airway inflammation in asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L300–L307 (2012).
Eleftheriadis, T. et al. Dichloroacetate at therapeutic concentration alters glucose metabolism and induces regulatory T-cell differentiation in alloreactive human lymphocytes. J. Basic Clin. Physiol. Pharmacol. 24, 271–276 (2013).
Thomas, S. et al. Methotrexate is a JAK/STAT pathway inhibitor. PLoS ONE 10, e0130078 (2015).
Shuvalov, O. et al. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget http://dx.doi.org/10.18632/oncotarget.15053 (2017).
Yin, Y. et al. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl Med. 7, 274ra18 (2015).
Sener, Z., Cederkvist, F. H., Volchenkov, R., Holen, H. L. & Skalhegg, B. S. T helper cell activation and expansion is sensitive to glutaminase inhibition under both hypoxic and normoxic conditions. PLoS ONE 11, e0160291 (2016).
Lee, C. F. et al. Preventing allograft rejection by targeting immune metabolism. Cell Rep. 13, 760–770 (2015).
Johnson, K. M. et al. Identification and validation of the mitochondrial F1F0-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem. Biol. 12, 485–496 (2005).
Gatza, E. et al. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Sci. Transl Med. 3, 67ra8 (2011).
Bednarski, J. J. et al. Attenuation of autoimmune disease in Fas-deficient mice by treatment with a cytotoxic benzodiazepine. Arthritis Rheum. 48, 757–766 (2003).
Buskiewicz, I. A. et al. Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci. Signal. 9, ra115 (2016).
Hua, S. & Dias, T. H. Hypoxia-inducible factor (HIF) as a target for novel therapies in rheumatoid arthritis. Front. Pharmacol. 7, 184 (2016).
Telang, S. et al. Small molecule inhibition of 6-phosphofructo-2-kinase suppresses T cell activation. J. Transl Med. 10, 95 (2012).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Acknowledgements
The authors would like to thank members of the Rathmell and Major labs for their contributions and intellectual input. The authors' work is supported by the Alliance for Lupus Research (J.C.R.), NIH National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK105550 (J.C.R.), the Lupus Research Alliance (A.S.M.), U.S. Department of Veterans Affairs Merit Award I0BX002968 (A.S.M.) and NIH National Heart, Lung, and Blood Institute grant F31 HL128040 (J.P.R.).
Author information
Authors and Affiliations
Contributions
All authors researched the data for the article, provided a substantial contribution to discussions of the content and contributed to writing the article and to review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Rhoads, J., Major, A. & Rathmell, J. Fine tuning of immunometabolism for the treatment of rheumatic diseases. Nat Rev Rheumatol 13, 313–320 (2017). https://doi.org/10.1038/nrrheum.2017.54
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2017.54
This article is cited by
-
Implications of immunometabolism for smouldering MS pathology and therapy
Nature Reviews Neurology (2023)
-
Impaired mitochondria of Tregs decreases OXPHOS-derived ATP in primary immune thrombocytopenia with positive plasma pathogens detected by metagenomic sequencing
Experimental Hematology & Oncology (2022)
-
Increase of aerobic glycolysis mediated by activated T helper cells drives synovial fibroblasts towards an inflammatory phenotype: new targets for therapy?
Arthritis Research & Therapy (2021)
-
Novel standardized method for extracellular flux analysis of oxidative and glycolytic metabolism in peripheral blood mononuclear cells
Scientific Reports (2021)
-
Lactate Production Precedes Inflammatory Cell Recruitment in Arthritic Ankles: an Imaging Study
Molecular Imaging and Biology (2020)