Key Points
-
The highly polygenic aetiology of rheumatoid arthritis (RA) is well characterized in population genetic studies, with >100 susceptibility loci reported
-
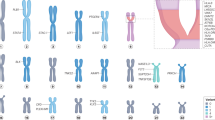
Genetic variants affecting the structure of epitope-binding sites of HLA molecules exhibit the strongest contributions to RA development; log-additive and non-log-additive effects of HLA alleles explain HLA–RA associations
-
The majority of non-HLA associations are mapped at noncoding variants that colocalize with cell-type-specific characteristics (for example, histone markers or enhancers) in CD4+ T cells
-
Pinpointing a single causal gene in a locus that contains multiple genes usually requires additional 'omics' data
-
RA-risk loci generally contain genes related to T-cell and B-cell pathways, cytokine signalling pathways, proliferation and/or impaired haematopoietic and immune systems
-
Many proteins encoded within RA-risk loci, and their interaction partners, are the targets of currently approved therapies, implicating a potential guiding role for human RA genetics data in drug discovery and repurposing
Abstract
Human genetic studies into rheumatoid arthritis (RA) have uncovered more than 100 genetic loci associated with susceptibility to RA and have refined the RA-association model for HLA variants. The majority of RA-risk variants are highly shared across multiple ancestral populations and are located in noncoding elements that might have allele-specific regulatory effects in relevant tissues. Emerging multi-omics data, high-density genotype data and bioinformatic approaches are enabling researchers to use RA-risk variants to identify functionally relevant cell types and biological pathways that are involved in impaired immune processes and disease phenotypes. This Review summarizes reported RA-risk loci and the latest insights from human genetic studies into RA pathogenesis, including how genetic data has helped to identify currently available drugs that could be repurposed for patients with RA and the role of genetics in guiding the development of new drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Aho, K., Koskenvuo, M., Tuominen, J. & Kaprio, J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 13, 899–902 (1986).
Silman, A. J. et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol. 32, 903–907 (1993).
MacGregor, A. J. et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 43, 30–37 (2000).
van der Woude, D. et al. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 60, 916–923 (2009).
Terao, C. et al. A twin study of rheumatoid arthritis in the Japanese population. Mod. Rheumatol. 26, 685–689 (2016).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Lenz, T. L. et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 47, 1085–1090 (2015).
Stahl, E. A. et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44, 483–489 (2012).
Horton, R. et al. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 889–899 (2004).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987).
Bang, S. Y., Lee, H. S., Lee, K. W. & Bae, S. C. Interaction of HLA-DRB1*09:01 and *04:05 with smoking suggests distinctive mechanisms of rheumatoid arthritis susceptibility beyond the shared epitope. J. Rheumatol. 40, 1054–1062 (2013).
Shimane, K. et al. An association analysis of HLA-DRB1 with systemic lupus erythematosus and rheumatoid arthritis in a Japanese population: effects of *09:01 allele on disease phenotypes. Rheumatology (Oxford) 52, 1172–1182 (2013).
Reynolds, R. J. et al. HLA-DRB1-associated rheumatoid arthritis risk at multiple levels in African Americans: hierarchical classification systems, amino acid positions, and residues. Arthritis Rheumatol. 66, 3274–3282 (2014).
Lee, H. S. et al. Increased susceptibility to rheumatoid arthritis in Koreans heterozygous for HLA-DRB1*0405 and *0901. Arthritis Rheum. 50, 3468–3475 (2004).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Kim, K. et al. Imputing variants in HLA-DR beta genes reveals that HLA-DRB1 is solely associated with rheumatoid arthritis and systemic lupus erythematosus. PLoS ONE 11, e0150283 (2016).
Okada, Y. et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 23, 6916–6926 (2014).
Han, B. et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am. J. Hum. Genet. 94, 522–532 (2014).
Anderson, K. M. et al. A molecular analysis of the shared epitope hypothesis binding of arthritogenic peptides to DRB1*04 alleles. Arthritis Rheumatol. 68, 1627–1636 (2016).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015).
Kim, K. et al. Interactions between amino acid-defined major histocompatibility complex class II variants and smoking in seropositive rheumatoid arthritis. Arthritis Rheumatol. 67, 2611–2623 (2015).
Kurreeman, F. et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am. J. Hum. Genet. 88, 57–69 (2011).
Terao, C. et al. A large-scale association study identified multiple HLA-DRB1 alleles associated with ACPA-negative rheumatoid arthritis in Japanese subjects. Ann. Rheum. Dis. 70, 2134–2139 (2011).
Viatte, S. et al. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann. Rheum. Dis. 71, 1984–1990 (2012).
Bossini-Castillo, L. et al. A genome-wide association study of rheumatoid arthritis without antibodies against citrullinated peptides. Ann. Rheum. Dis. 74, e15 (2015).
Wagner, C. A. et al. Identification of anticitrullinated protein antibody reactivities in a subset of anti-CCP-negative rheumatoid arthritis: association with cigarette smoking and HLA-DRB1 'shared epitope' alleles. Ann. Rheum. Dis. 74, 579–586 (2015).
Kim, K. et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann. Rheum. Dis. 74, e13 (2015).
Cobb, J. E. et al. Identification of the tyrosine-protein phosphatase non-receptor type 2 as a rheumatoid arthritis susceptibility locus in Europeans. PLoS ONE 8, e66456 (2013).
McAllister, K. et al. Identification of BACH2 and RAD51B as rheumatoid arthritis susceptibility loci in a meta-analysis of genome-wide data. Arthritis Rheum. 65, 3058–3062 (2013).
Orozco, G. et al. Novel rheumatoid arthritis susceptibility locus at 22q12 identified in an extended UK genome-wide association study. Arthritis Rheumatol. 66, 24–30 (2014).
Okada, Y. et al. Significant impact of miRNA-target gene networks on genetics of human complex traits. Sci. Rep. 6, 22223 (2016).
Kim, K. et al. Association-heterogeneity mapping identifies and Asian-spcific association of the GTF2I locus with rheumatoid arthritis. Sci. Rep. 6, 27563 (2016).
Fortune, M. D. et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015).
Gusev, A. et al. Quantifying missing heritability at known GWAS loci. PLoS Genet. 9, e1003993 (2013).
Okada, Y. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 44, 511–516 (2012).
Kurreeman, F. A. et al. Use of a multiethnic approach to identify rheumatoid- arthritis-susceptibility loci, 1p36 and 17q12. Am. J. Hum. Genet. 90, 524–532 (2012).
Farh, K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Kichaev, G. & Pasaniuc, B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97, 260–271 (2015).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Trynka, G. et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 97, 139–152 (2015).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Freudenberg, J., Gregersen, P. & Li, W. Enrichment of genetic variants for rheumatoid arthritis within T-cell and NK-cell enhancer regions. Mol. Med. 21, 180–184 (2015).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Hrdlickova, B. et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 6, 88 (2014).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Carpenter, S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013).
Hyatt, G. et al. Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol. 7, 686–691 (2006).
Hu, X. et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet. 89, 496–506 (2011).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Hu, X. et al. Regulation of gene expression in autoimmune disease loci and the genetic basis of proliferation in CD4+ effector memory T cells. PLoS Genet. 10, e1004404 (2014).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Stratigopoulos, G., LeDuc, C. A., Cremona, M. L., Chung, W. K. & Leibel, R. L. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J. Biol. Chem. 286, 2155–2170 (2011).
Stratigopoulos, G. et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 19, 767–779 (2014).
Martin, P. et al. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 6, 10069 (2015).
Segre, A. V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Eppig, J. T. et al. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 40, D881–D886 (2012).
de Rooy, D. P. et al. Genetic studies on components of the Wnt signalling pathway and the severity of joint destruction in rheumatoid arthritis. Ann. Rheum. Dis. 72, 769–775 (2013).
Yarwood, A. et al. Enrichment of vitamin D response elements in RA-associated loci supports a role for vitamin D in the pathogenesis of RA. Genes Immun. 14, 325–329 (2013).
Liu, G. et al. Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. PLoS ONE 8, e75951 (2013).
Andreassen, O. A. et al. Abundant genetic overlap between blood lipids and immune-mediated diseases indicates shared molecular genetic mechanisms. PLoS ONE 10, e0123057 (2015).
Buchel, F. et al. Integrative pathway-based approach for genome-wide association studies: identification of new pathways for rheumatoid arthritis and type 1 diabetes. PLoS ONE 8, e78577 (2013).
Maggadottir, S. M. & Sullivan, K. E. The intersection of immune deficiency and autoimmunity. Curr. Opin. Rheumatol. 26, 570–578 (2014).
Timmons, J. A., Szkop, K. J. & Gallagher, I. J. Multiple sources of bias confound functional enrichment analysis of global-omics data. Genome Biol. 16, 186 (2015).
Atzeni, F. et al. Different effects of biological drugs in rheumatoid arthritis. Autoimmun. Rev. 12, 575–579 (2013).
Grover, M. P. et al. Identification of novel therapeutics for complex diseases from genome-wide association data. BMC Med. Genomics 7 (Suppl. 1), S8 (2014).
Maloney, D. G. et al. IDEC-C2B8 (rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin's lymphoma. Blood 90, 2188–2195 (1997).
Edwards, J. C. et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N. Engl. J. Med. 350, 2572–2581 (2004).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Li, G. et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 9, e1003487 (2013).
Ekwall, A. K. et al. The rheumatoid arthritis risk gene LBH regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 67, 1193–1202 (2015).
Giles, J. L., Choy, E., van den Berg, C., Morgan, B. P. & Harris, C. L. Functional analysis of a complement polymorphism (rs17611) associated with rheumatoid arthritis. J. Immunol. 194, 3029–3034 (2015).
Schuster, C. et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity 42, 942–952 (2015).
Chang, H. H., Dwivedi, N., Nicholas, A. P. & Ho, I. C. The W620 polymorphism in PTPN22 disrupts its interaction with peptidylarginine deiminase type 4 and enhances citrullination and NETosis. Arthritis Rheumatol. 67, 2323–2334 (2015).
Spurlock, C. F. III, Tossberg, J. T., Olsen, N. J. & Aune, T. M. Cutting edge: chronic NF-κB activation in CD4+ T Cells in rheumatoid arthritis is genetically determined by HLA risk alleles. J. Immunol. 195, 791–795 (2015).
Muthana, M. et al. C5orf30 is a negative regulator of tissue damage in rheumatoid arthritis. Proc. Natl Acad. Sci. USA 112, 11618–11623 (2015).
Messemaker, T. C. et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 17, 85–92 (2015).
de la Puerta, M. L. et al. The autoimmunity risk variant LYP-W620 cooperates with CSK in the regulation of TCR signaling. PLoS ONE 8, e54569 (2013).
Han, T. U. et al. Association of an activity-enhancing variant of IRAK1 and an MECP2-IRAK1 haplotype with increased susceptibility to rheumatoid arthritis. Arthritis Rheum. 65, 590–598 (2013).
Ferreira, R. C. et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 9, e1003444 (2013).
Lewis, M. J. et al. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 96, 221–234 (2015).
Suzuki, A. & Yamamoto, K. From genetics to functional insights into rheumatoid arthritis. Clin. Exp. Rheumatol. 33 (4 Suppl. 92), S40–S43 (2015).
Viatte, S. et al. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA 313, 1645–1656 (2015).
Visscher, P. M., Hill, W. G. & Wray, N. R. Heritability in the genomics era — concepts and misconceptions. Nat. Rev. Genet. 9, 255–266 (2008).
Svendsen, A. J. et al. On the origin of rheumatoid arthritis: the impact of environment and genes — a population based twin study. PLoS ONE 8, e57304 (2013).
Diogo, D. et al. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am. J. Hum. Genet. 92, 15–27 (2013).
Diogo, D. et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS ONE 10, e0122271 (2015).
Bang, S. Y. et al. Targeted exon sequencing fails to identify rare coding variants with large effect in rheumatoid arthritis. Arthritis Res. Ther. 16, 447 (2014).
Okada, Y. et al. Integration of sequence data from a consanguineous family with genetic data from an outbred population identifies PLB1 as a candidate rheumatoid arthritis risk gene. PLoS ONE 9, e87645 (2014).
Veal, C. D. et al. A 129-kb deletion on chromosome 12 confers substantial protection against rheumatoid arthritis, implicating the gene SLC2A3. Hum. Mutat. 35, 248–256 (2014).
Du, Y. et al. Contribution of functional LILRA3, but not nonfunctional LILRA3, to sex bias in susceptibility and severity of anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol. 66, 822–830 (2014).
Chen, J. Y. et al. Association of FCGR3A and FCGR3B copy number variations with systemic lupus erythematosus and rheumatoid arthritis in Taiwanese patients. Arthritis Rheumatol. 66, 3113–3121 (2014).
Franke, L. et al. Association analysis of copy numbers of FC-gamma receptor genes for rheumatoid arthritis and other immune-mediated phenotypes. Eur. J. Hum. Genet. 24, 263–270 (2016).
Wu, C. C. et al. Whole-genome detection of disease-associated deletions or excess homozygosity in a case–control study of rheumatoid arthritis. Hum. Mol. Genet. 22, 1249–1261 (2013).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Acknowledgements
K.K. and S.C.B. are funded by the Korea Healthcare Technology R&D Project from the Ministry for Health and Welfare, Republic of Korea, grant numbers HI15C3182 (K.K) and HI13C2124 (S.C.B.).
Author information
Authors and Affiliations
Contributions
K.K. and S.C.B. collected data for the article and wrote the manuscript. All authors provided substantial contributions to the discussion of content and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
Supplementary information S1 (figure)
Reported rheumatoid arthritis (RA) loci. (PDF 808 kb)
Rights and permissions
About this article
Cite this article
Kim, K., Bang, SY., Lee, HS. et al. Update on the genetic architecture of rheumatoid arthritis. Nat Rev Rheumatol 13, 13–24 (2017). https://doi.org/10.1038/nrrheum.2016.176
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2016.176
This article is cited by
-
Association of HLA-DRB1 locus with treatment response to abatacept or TNF inhibitors in patients with seropositive rheumatoid arthritis
Scientific Reports (2024)
-
A Common Functional Variant at the Enhancer of the Rheumatoid Arthritis Risk Gene ORMDL3 Regulates its Expression Through Allele-Specific JunD Binding
Phenomics (2023)
-
The genome editing revolution: review
Journal of Genetic Engineering and Biotechnology (2020)
-
T helper cells in synovial fluid of patients with rheumatoid arthritis primarily have a Th1 and a CXCR3+Th2 phenotype
Arthritis Research & Therapy (2020)
-
Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes
Nature Reviews Rheumatology (2020)