Key Points
-
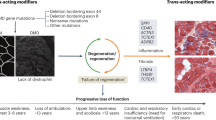
New sequencing technologies, including whole-exome and whole-genome sequencing, combined with increased data sharing and international research collaborations, are resulting in the identification of new limb-girdle muscular dystrophy (LGMD) subtypes and diagnosis of additional patients
-
These technologies are also providing evidence that added complexities, including intronic and regulatory mutations, oligogenic inheritance and modifier gene effects, could have a role in the hardest-to-diagnose forms of LGMD
-
The increased molecular and pathogenetic understanding of LGMD subtypes is now calling into question their original classification by phenotype, and new systems-based and pathway-based classification systems may be required
-
'Trial readiness', which includes preparation of patient cohorts, care standards, outcome measures, and biomarkers stratified according to genes and/or pathways, is a key concept for translation of basic research to the clinic
-
Collaboration between patients, industry and academia is essential in the translational pathway towards therapy development and clinical trials
Abstract
The limb-girdle muscular dystrophies (LGMDs) are a diverse group of genetic neuromuscular conditions that usually manifest in the proximal muscles of the hip and shoulder girdles. Since the identification of the first gene associated with the phenotype in 1994, an extensive body of research has identified the genetic defects responsible for over 30 LGMD subtypes, revealed an increasingly varied phenotypic spectrum, and exposed the need to move towards a systems-based understanding of the molecular pathways affected. New sequencing technologies, including whole-exome and whole-genome sequencing, are continuing to expand the range of genes and phenotypes associated with the LGMDs, and new computational approaches are helping clinicians to adapt to this new genomic medicine paradigm. However, 60 years on from the first description of LGMD, no curative therapies exist, and systematic exploration of the natural history is still lacking. To enable rapid translation of basic research to the clinic, well-phenotyped and genetically characterized patient cohorts are a necessity, and appropriate outcome measures and biomarkers must be developed through natural history studies. Here, we review the international collaborations that are addressing these translational research issues, and the lessons learned from large-scale LGMD sequencing programmes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Walton, J. N. & Nattrass, F. J. On the classification, natural history and treatment of the myopathies. Brain 77, 169–231 (1954).
Fanin, M., Nascimbeni, A. C., Fulizio, L. & Angelini, C. The frequency of limb girdle muscular dystrophy 2A in northeastern Italy. Neuromuscul. Disord. 15, 218–224 (2005).
Narayanaswami, P. et al. Evidence-based guideline summary: diagnosis and treatment of limb-girdle and distal dystrophies: report of the Guideline Development Subcommittee of the American Academy of Neurology and the Practice Issues Review Panel of the American Association of Neuromuscular and Electrodiagnostic Medicine. Neurology 83, 1453–1463 (2014).
Nigro, V. & Savarese, M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 33, 1–12 (2014).
Murphy, A. P. & Straub, V. The classification, natural history and treatment of the limb girdle muscular dystrophies. J. Neuromuscul. Dis. 2 (Suppl. 2), 7–19 (2015).
Hauser, M. A. et al. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 9, 2141–2147 (2000).
Muchir, A. et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 9, 1453–1459 (2000).
Minetti, C. et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat. Genet. 18, 365–368 (1998).
McNally, E. Caveolin-3 in muscular dystrophy. Hum. Mol. Genet. 7, 871–877 (1998).
Sarparanta, J. et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 44, 450–455 (2012).
Harms, M. B. et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol. 71, 407–416 (2012).
Greenberg, S. A. et al. Etiology of limb girdle muscular dystrophy 1D/1E determined by laser capture microdissection proteomics. Ann. Neurol. 71, 141–145 (2012).
Hedberg, C., Melberg, A., Kuhl, A., Jenne, D. & Oldfors, A. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy 7 is caused by a DES mutation. Eur. J. Hum. Genet. 20, 984–985 (2012).
Melia, M. J. et al. Limb-girdle muscular dystrophy 1F is caused by a microdeletion in the transportin 3 gene. Brain 136, 1508–1517 (2013).
Vieira, N. M. et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum. Mol. Genet. 23, 4103–4110 (2014).
Beckmann, J. S. et al. A gene for limb-girdle muscular dystrophy maps to chromosome 15 by linkage. C. R. Acad. Sci. III 312, 141–148 (1991).
Richard, I. et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27–40 (1995).
Bashlr, R. et al. A gene for autosomal recessive limb-girdle muscular dystrophy maps to chromosome 2p. Hum. Mol. Genet. 3, 455–457 (1994).
Liu, J. et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20, 31–36 (1998).
Bashir, R. et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 20, 37–42 (1998).
Azlbi, K. et al. Severe childhood autosomal recessive muscular dystrophy with the deficiency of the 50 kDa dystrophin-associated glycoprotein maps to chromosome 13q12. Hum. Mol. Genet. 2, 1423–1428 (1993).
Noguchi, S. et al. Mutations in the dystrophin-associated protein γ-sarcoglycan in chromosome 13 muscular dystrophy. Science 270, 819–822 (1995).
Roberds, S. L. et al. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell 78, 625–633 (1994).
Piccolo, F. et al. Primary adhalinopathy: a common cause of autosomal recessive muscular dystrophy of variable severity. Nat. Genet. 10, 243–245 (1995).
Lim, L. E. et al. β-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat. Genet. 11, 257–265 (1995).
Bönnemann, C. G. et al. β-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 11, 266–273 (1995).
Passos-Bueno, M. Linkage analysis in autosomal recessive limb-girdle muscular dystrophy (AR LGMD) maps a sixth form to 5q33–34 (LGMD2F) and indicates that there is at least one more subtype of AR LGMD. Hum. Mol. Genet. 5, 815–820 (1996).
Nigro, V. et al. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the δ-sarcoglycan gene. Nat. Genet. 14, 195–198 (1996).
Moreira, E. S. et al. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat. Genet. 24, 163–166 (2000).
Weiler, T. et al. A gene for autosomal recessive limb-girdle muscular dystrophy in Manitoba Hutterites maps to chromosome region 9q31–q33: evidence for another limb-girdle muscular dystrophy locus. Am. J. Hum. Genet. 63, 140–147 (1998).
Frosk, P. et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 70, 663–672 (2002).
Brockington, M. et al. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum. Mol. Genet. 10, 2851–2859 (2001).
Hackman, P. et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 71, 492–500 (2002).
Balci, B. et al. An autosomal recessive limb girdle muscular dystrophy (LGMD2) with mild mental retardation is allelic to Walker–Warburg syndrome (WWS) caused by a mutation in the POMT1 gene. Neuromuscul. Disord. 15, 271–275 (2005).
Jarry, J. et al. A novel autosomal recessive limb-girdle muscular dystrophy with quadriceps atrophy maps to 11p13–p12. Brain 130, 368–380 (2007).
Bolduc, V. et al. Recessive mutations in the putative calcium-activated chloride channel anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86, 213–221 (2010).
Murakami, T. et al. Fukutin gene mutations cause dilated cardiomyopathy with minimal muscle weakness. Ann. Neurol. 60, 597–602 (2006).
Godfrey, C. et al. Fukutin gene mutations in steroid-responsive limb girdle muscular dystrophy. Ann. Neurol. 60, 603–610 (2006).
Biancheri, R. et al. POMT2 gene mutation in limb-girdle muscular dystrophy with inflammatory changes. Biochem. Biophys. Res. Commun. 363, 1033–1037 (2007).
Clement, E. M. et al. Mild POMGnT1 mutations underlie a novel limb-girdle muscular dystrophy variant. Arch. Neurol. 65, 137–141 (2008).
Raducu, M., Baets, J., Fano, O., Van Coster, R. & Cruces, J. Promoter alteration causes transcriptional repression of the POMGNT1 gene in limb-girdle muscular dystrophy type 2O. Eur. J. Hum. Genet. 20, 945–952 (2012).
Hara, Y. et al. A dystroglycan mutation associated with limb-girdle muscular dystrophy. N. Engl. J. Med. 364, 939–946 (2011).
Gundesli, H. et al. Mutation in exon 1f of PLEC, leading to disruption of plectin isoform 1f, causes autosomal-recessive limb-girdle muscular dystrophy. Am. J. Hum. Genet. 87, 834–841 (2010).
Cetin, N. et al. A novel desmin mutation leading to autosomal recessive limb-girdle muscular dystrophy: distinct histopathological outcomes compared with desminopathies. J. Med. Genet. 50, 437–443 (2013).
Bögershausen, N. et al. Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am. J. Hum. Genet. 93, 181–190 (2013).
Carss, K. J. et al. Mutations in GDP-mannose pyrophosphorylase b cause congenital and limb-girdle muscular dystrophies associated with hypoglycosylation of α-dystroglycan. Am. J. Hum. Genet. 93, 29–41 (2013).
Tasca, G. et al. Limb-girdle muscular dystrophy with α-dystroglycan deficiency and mutations in the ISPD gene. Neurology 80, 963–965 (2013).
Preisler, N. et al. Late-onset Pompe disease is prevalent in unclassified limb-girdle muscular dystrophies. Mol. Genet. Metab. 110, 287–289 (2013).
Chardon, J. W. et al. LIMS2 mutations are associated with a novel muscular dystrophy, severe cardiomyopathy and triangular tongues. Clin. Genet. 88, 558–5564 (2015).
Schindler, R. F. et al. POPDC1S201F causes muscular dystrophy and arrhythmia by affecting protein trafficking. J. Clin. Invest. 126, 239–253 (2016).
Wells, L. The O-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J. Biol. Chem. 288, 6930–6935 (2013).
Endo, T. Glycobiology of α-dystroglycan and muscular dystrophy. J. Biochem. 157, 1–12 (2015).
Yoshida-Moriguchi, T. & Campbell, K. P. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology 25, 702–713 (2015).
Vissing, J., Sveen, M. L. & Duno, M. O. 22 dominant inheritance of limb girdle muscular dystrophy type 2A. Neuromuscul. Disord. 21, 750 (2011).
Ghaoui, R. et al. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: outcomes and lessons learned. JAMA Neurol. 72, 1424–1432 (2015).
Biancalana, V. & Laporte, J. Diagnostic use of massively parallel sequencing in neuromuscular diseases: towards an integrated diagnosis. J. Neuromuscul. Dis. 2, 193–203 (2015).
Chae, J. H. et al. Utility of next generation sequencing in genetic diagnosis of early onset neuromuscular disorders. J. Med. Genet. 52, 208–216 (2015).
Ankala, A. et al. A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann. Neurol. 77, 206–214 (2014).
Cabrera-Serrano, M. et al. Expanding the phenotype of GMPPB mutations. Brain 138, 836–844 (2015).
Lek, M. & MacArthur, D. The challenge of next generation sequencing in the context of neuromuscular diseases. J. Neuromuscul. Dis. 1, 135–149 (2014).
Boycott, K. M., Dyment, D. A., Sawyer, S. L., Vanstone, M. R. & Beaulieu, C. L. Identification of genes for childhood heritable diseases. Annu. Rev. Med. 65, 19–31 (2014).
Rodger, S. et al. The TREAT-NMD care and trial site registry: an online registry to facilitate clinical research for neuromuscular diseases. Orphanet J. Rare Dis. 8, 171 (2013).
Bushby, K., Lynn, S. & Straub, T. Collaborating to bring new therapies to the patient — the TREAT-NMD model. Acta Myol. 28, 12–15 (2009).
Knoppers, B. M., Harris, J. R., Budin- Ljøsne, I. & Dove, E. S. A human rights approach to an international code of conduct for genomic and clinical data sharing. Hum. Genet. 133, 895–903 (2014).
Lappalainen, I. et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 47, 692–695 (2015).
Thompson, R. et al. RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J. Gen. Intern. Med. 29, S780–S787 (2014).
Monaco, L., Crimi, M. & Wang, C. M. The challenge for a European network of biobanks for rare diseases taken up by RD-Connect. Pathobiology 81, 231–236 (2014).
MacArthur, D. G. et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476 (2014).
Di Fruscio, G., Garofalo, A., Mutarelli, M., Savarese, M. & Nigro, V. Are all the previously reported genetic variants in limb girdle muscular dystrophy genes pathogenic? Eur. J. Hum. Genet. 24, 73–77 (2015).
Norwood, F. L. et al. Prevalence of genetic muscle disease in Northern England: in-depth analysis of a muscle clinic population. Brain 132, 3175–3186 (2009).
Köhler, S. et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 42, D966–D974 (2014).
Philippakis, A. A. et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 36, 915–921 (2015).
Faravelli, I., Nizzardo, M., Comi, G. P. & Corti, S. Spinal muscular atrophy — recent therapeutic advances for an old challenge. Nat. Rev. Neurol. 11, 351–359 (2015).
Jirka, S. & Aartsma-Rus, A. An update on RNA-targeting therapies for neuromuscular disorders. Curr. Opin. Neurol. 28, 515–521 (2015).
Klein, A. F., Dastidar, S., Furling, D. & Chuah, M. K. Therapeutic approaches for dominant muscle diseases: highlight on myotonic dystrophy. Curr. Gene Ther. 15, 329–337 (2015).
Straub, V. & Bertoli, M. Where do we stand in trial readiness for autosomal recessive limb girdle muscular dystrophies? Neuromuscul. Disord. 26, 111–125 (2016).
Bushby, K. Looking forward to new therapies: a personal perspective on the translational landscape for muscular dystrophies. J. Neuromuscul. Dis. 2, 83–87 (2015).
Bladen, C. L. et al. The TREAT-NMD Duchenne muscular dystrophy registries: conception, design, and utilization by industry and academia. Hum. Mutat. 34, 1449–1457 (2013).
Bladen, C. L. et al. Mapping the differences in care for 5,000 spinal muscular atrophy patients, a survey of 24 national registries in North America, Australasia and Europe. J. Neurol. 261, 152–163 (2013).
Walter, M. C. et al. Treatment of dysferlinopathy with deflazacort: a double-blind, placebo-controlled clinical trial. Orphanet J. Rare Dis. 8, 26 (2013).
Burch, P. M. et al. Muscle-derived proteins as serum biomarkers for monitoring disease progression in three forms of muscular dystrophy. J. Neuromuscul. Dis. 2, 241–255 (2015).
Mascalzoni, D. et al. International Charter of principles for sharing bio-specimens and data. Eur. J. Hum. Genet. 23, 721–728 (2014).
Acknowledgements
The work leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreements 305444 (RD-Connect) and 305121 (Neuromics). TREAT-NMD was funded under EU FP6 contract no. 036825 (2007–2011) and TREAT-NMD Operating Grant — Second Public Health Programme (contract no. 2012 3307). Funding has also been received from the Jain Foundation, the LGMD2I Research Fund, Genzyme and Ultragenyx Pharmaceutical.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all stages of the preparation of this article.
Corresponding author
Ethics declarations
Competing interests
V.S. is or has been a principal investigator for trials sponsored by Genzyme, GSK, Prosensa/Biomarin, ISIS Pharmaceuticals, and Sarepta. He has received speaker honoraria from Genzyme and has been a member of the company's international Pompe advisory board. He also is or has been on advisory boards for Acceleron Pharma, Audentes Therapeutics, Bristol-Myers Squibb, Italfarmaco, Nicox, Pfizer, Prosensa, Santhera, Summit Therapeutics and TrophyNOD. He has research collaborations with Ultragenyx pharmaceutical and Genzyme/Sanofi. None of the industry collaborations pose a conflict of interest for this Review article. R.T. declares no competing interests.
Rights and permissions
About this article
Cite this article
Thompson, R., Straub, V. Limb-girdle muscular dystrophies — international collaborations for translational research. Nat Rev Neurol 12, 294–309 (2016). https://doi.org/10.1038/nrneurol.2016.35
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneurol.2016.35
This article is cited by
-
Gene therapy with bidridistrogene xeboparvovec for limb-girdle muscular dystrophy type 2E/R4: phase 1/2 trial results
Nature Medicine (2024)
-
Heterozygous missense variant in the TTN gene causing Tibial muscular dystrophy
Egyptian Journal of Medical Human Genetics (2022)
-
Reporting a rare form of myopathy, myopathy with extrapyramidal signs, in an Iranian family using next generation sequencing: a case report
BMC Medical Genetics (2020)
-
The unified myofibrillar matrix for force generation in muscle
Nature Communications (2020)
-
Identification of thiostrepton as a pharmacological approach to rescue misfolded alpha-sarcoglycan mutant proteins from degradation
Scientific Reports (2019)