Abstract
Hereditary amyotrophic lateral sclerosis (ALS) encompasses a group of genetic disorders characterized by adult-onset loss of the lower and upper motor neuron systems, often with involvement of other parts of the nervous system. Cases of hereditary ALS have been attributed to mutations in 12 different genes, the most common being SOD1, FUS and TARDBP—mutations in the other genes are rare. The identified genes explain 25–35% of cases of familial ALS, but identifying the remaining genes has proved difficult. Only a few genes seem to account for significant numbers of ALS cases, with many others causing a few cases each. Hereditary ALS can be inherited in an autosomal dominant, autosomal recessive or X-linked manner, and families with low disease penetrance are frequently observed. In such families, the genetic predisposition may remain unnoticed, so many patients carry a diagnosis of isolated or sporadic ALS. The only clinical feature that distinguishes recognized hereditary from apparently sporadic ALS is a lower mean age of onset in the former. All the clinical features reported in hereditary cases (including signs of extrapyramidal, cerebellar or cognitive involvement) have also been observed in sporadic cases. Genetic counseling and risk assessment in relatives depend on establishing the specific gene defect and the disease penetrance in the particular family.
Key Points
-
Familial amyotrophic lateral sclerosis (ALS) is frequently underdiagnosed, and apparently sporadic ALS may, in many cases, be familial ALS with reduced disease penetrance
-
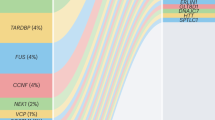
A few genes are associated with a substantial proportion of ALS cases, and many others probably contribute to only a few cases
-
Mutations in 12 genes have been found to cause familial ALS, the most common being SOD1, followed by FUS and TARDBP
-
All genes mutated in familial ALS have also been found mutated in patients diagnosed with sporadic ALS and, besides a lower mean age of onset, no clinical difference exists between the two groups
-
No ALS gene has exclusively been associated with an ALS-only motor phenotype, suggesting that ALS is a multisystem neurodegenerative syndrome with a propensity for targeting the motor system
-
Most ALS cases are probably attributable to oligogenic inheritance, perhaps in combination with environmental factors, but monogenic inheritance with a mutation private to each individual is also possible
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Aran, F. A. Research on an as yet undescribed disease of the muscular system (progressive muscular atrophy) [French]. Arch. Gén. Méd. 24, 15–35 (1848).
Charcot J. M. Lectures on the Diseases of the Nervous System, vol. 2, series 2 (ed. Sigerson, G.) 163–204 (New Sydenham Society, London, 1881).
Chio, A. et al. Large proportion of amyotrophic lateral sclerosis cases in Sardinia due to a single founder mutation of the TARDBP gene. Arch. Neurol. 68, 594–598 (2011).
van Es, M. A. et al. Large scale SOD1 mutation screening provides evidence for genetic heterogeneity in ALS. J. Neurol. Neurosurg. Psychiatry 81, 562–566 (2010).
Eisen, A. et al. SOD1 gene mutations in ALS patients in British Columbia, Canada: clinical features, neurophysiology and ethical issues in management. Amyotroph. Lateral Scler. 9, 108–119 (2008).
Andersen, P. M. et al. Phenotypic heterogeneity in MND-patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain 10, 1723–1737 (1997).
Li, T. M., Alberman, E. & Swash, M. Comparison of sporadic and familial disease amongst 580 cases of motor neuron disease. J. Neurol. Neurosurg. Psychiatry 51, 778–784 (1988).
Kurland, K. T. & Mulder, D. W. Epidemiological investigations of amyotrophic lateral sclerosis. Familial aggregations indicative of dominant inheritance. Part I & II. Neurology 5, 182–267 (1955).
Fallis, B. A. & Hardiman, O. Aggregation of neurodegenerative disease in ALS kindreds. Amyotroph. Lateral Scler. 10, 95–98 (2009).
Forsberg, K. et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PloS ONE 5, e11552 (2010).
Bosco, D. A. et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 13, 1396–1403 (2010).
Sojka, P., Andersen, P. M. & Forsgren, L. Effects of riluzole on symptom progression in amyotrophic lateral sclerosis. Lancet 349, 176–177 (1997).
Tikka, T. et al. Minocycline prevents neurotoxicity induced by cerebrospinal fluid from patients with motor neuron disease. Brain 125, 722–731 (2002).
Swash, M. & Leigh, N. Criteria for diagnosis of familial amyotrophic lateral sclerosis. European FALS Collaborative Group. Neuromuscul. Disord. 2, 7–9 (1992).
Stewart, H. G. & Andersen, P. M. Neurophysiology of hereditary ALS, in Clinical Neurophysiology of Motor Neuron Diseases (ed. Eisen, A.) 543–562 (Elsevier, Amsterdam, 2004).
Byrne, S. et al. Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 82, 623–627 (2010).
Byrne, S. et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 12, 157–159 (2011).
Johnston, C. A. et al. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J. Neurol. 253, 1642–1643 (2006).
Hand, C. K. et al. Compound heterozygous D90A and D96N SOD1 mutations in a recessive amyotrophic lateral sclerosis family. Ann. Neurol. 49, 267–271 (2001).
DeJesus-Hernandez, M. et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 31, 1377–1389 (2010).
Chio, A. et al. A de novo missense mutation of the FUS gene in a “true” sporadic ALS case. Neurobiol. Aging 32, 553.e23–553.e25 (2011).
Al-Chalabi, A. & Lewis C. M. Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum. Hered. 71, 281–288 (2011).
Williams, D. B., Floate, D. A. & Leicester, J. Familial motor neuron disease: differing penetrance in large pedigrees. J. Neurol. Sci. 86, 215–230 (1988).
Jones, C. T., Swingler, R. J. & Brock, D. J. H. Identification of a novel SOD1 mutation in an apparantly sporadic amyotrophic lateral sclerosis patient and the detection of Ile113Thr in three others. Hum. Mol. Genet. 3, 649–650 (1994).
Cudkowicz, M. et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 41, 210–221 (1997).
Juneja, T., Pericak-Vance, M. A., Laing, N. G., Dave, S. & Siddique, T. Prognosis in familial amyotrophic lateral sclerosis: progression and survival in patients with glu100gly and ala4val mutations in Cu,Zn superoxide dismutase. Neurology 48, 55–57 (1997).
Andersen, P. M. et al. Autosomal recessive adult-onset ALS associated with homozygosity for Asp90Ala CuZn-superoxide dismutase mutation. A clinical and genealogical study of 36 patients. Brain 119, 1153–1172 (1996).
Millecamps, S. et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype–phenotype correlations. J. Med. Genet. 47, 554–560 (2010).
Wroe, R., Wai-Ling Butler, A., Andersen, P. M., Powell, J. F. & Al-Chalabi, A. ALSOD: the Amyotrophic Lateral Sclerosis Online Database. Amyotroph. Lateral Scler. 9, 249–250 (2008).
ALS Online Genetics database [online], (2011).
Rosen, D. R. et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993).
Andersen, P. M. et al. Sixteen novel mutations in the Cu/Zn superoxide dismutase genes in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2, 62–73 (2003).
Valdmanis, P. N. et al. A mutation that creates a pseudoexon in SOD1 causes familial ALS. Ann. Hum. Genet. 73, 652–657 (2009).
Zinman, L. et al. A mechanism for low penetrance in an ALS family with a novel SOD1 deletion. Neurology 72, 1153–1159 (2009).
Birve, A. et al. A novel SOD1 splice site mutation associated with familial ALS revealed by SOD activity analysis. Hum. Mol. Genet. 19, 4201–4206 (2010).
Brotherton, T. et al. A novel ALS SOD1 C6S mutation with implications for aggregation-related toxicity and genetic counseling. Amyotroph. Lateral Scler. 12, 215–219 (2011).
Andersen, P. M. et al. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat. Genet. 10, 61–66 (1995).
Prudencio, M., Hart, P. J., Borchelt, D. R. & Andersen, P. M. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum. Mol. Genet. 18, 3217–3226 (2009).
Gurney, M. E. et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994).
Reaume, A. G. et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 13, 43–47 (1996).
Ben Hamida, M., Hentati, F. & Ben Hamida, C. Hereditary motor system diseases (chronic juvenile amyotrophic lateral sclerosis). Conditions combining a bilateral pyramidal syndrome with limb and bulbar amyotrophy. Brain 113, 347–363 (1990).
Hadano, S. et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis. Nat. Genet. 29, 166–173 (2001).
Chen, Y. Z. et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74, 1128–1135 (2004).
Hirano, M. et al. Senataxin mutations and amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 12, 223–227 (2011).
Yasser, S. et al. An unusual case of familial ALS and cerebellar ataxia. Amyotroph. Lateral Scler. 11, 568–570 (2010).
Moreira, M. C. et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia–ocular apraxia 2. Nat. Genet. 36, 225–227 (2004).
Gitcho, M. A. et al. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 63, 535–538 (2008).
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Kabashi, E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 (2008).
Daoud, H. et al. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J. Med. Genet. 46, 112–114 (2009).
Kühlein, P. et al. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Arch. Neurol. 65, 1185–1189 (2008).
Kirby, J. et al. Broad clinical phenotypes associated with TAR-DNA binding protein (TARDBP) mutations in amyotrophic lateral sclerosis. Neurogenetics 11, 217–225 (2010).
Tsai, C.-P. et al. FUS, TARDBP and SOD1 mutations in Taiwanese cohort with familial ALS. Neurobiol. Aging 32, 553.e13–553.e21 (2011).
Origone, P. et al. Enlarging clinical spectrum of FALS with TARDBP gene mutations: S393L variant in an Italian family showing phenotypic variability and relevance for genetic counselling. Amyotroph. Lateral Scler. 11, 223–227 (2010).
Gitcho, M. A. et al. TARDBP 3'-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 118, 633–645 (2009).
Benajiba, L. et al. TARDBP mutations in motor neuron disease with frontotemporal lobar degeneration. Ann. Neurol. 65, 470–473 (2009).
Kovacs, G. G. et al. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov. Disord. 24, 1843–1847 (2009).
Borroni, B. et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 30, E974–E983 (2009).
Quadri, M. et al. Broadening the phenotype of TARDBP mutations: the TARDBP Ala382Thr mutation and Parkinson's disease in Sardinia. Neurogenetics 12, 203–209 (2011).
Vance, C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009).
Kwiatkowski, T. J. Jr et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009).
Ticozzi, N. et al. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology 73, 1180–1185 (2009).
Yan, J. et al. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology 75, 807–814 (2010).
Blair, I. P. et al. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J. Neurol. Neurosurg. Psychiatry 81, 639–645 (2010).
Van Langenhove, T. et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology 74, 366–371 (2010).
Nishimura, A. L. et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 75, 822–831 (2004).
Landers, J. E. et al. New VAPB deletion variant and exclusion of VAPB mutations in familial ALS. Neurology 70, 1179–1185 (2008).
Funke, A. D. et al. The p.P56S mutation in the VAPB gene is not due to a single founder: the first European case. Clin. Genet. 77, 302–303 (2010).
Chen, H. J. et al. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J. Biol. Chem. 285, 40266–40281 (2010).
Greenway, M. J. et al. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat. Genet. 38, 411–413 (2006).
van Es, M. A. et al. A case of ALS–FTD in a large FALS pedigree with a K17I ANG mutation. Neurology 72, 287–288 (2009).
Maruyama, H. et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 (2010).
Millecamps, S. et al. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiol. Aging. 32, e11–e13 (2011).
Belzil, V. V. et al. Analysis of OPTN as a causative gene for amyotrophic lateral sclerosis. Neurobiol. Aging. 32, e13–e14 (2011).
Elden, A. C. et al. Ataxin-2 intermediate length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010).
Lee, T. et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 20, 1697–1700 (2011).
Daoud, H. et al. Association of long ATXN2 CAG repeat sizes with increased risk of amyotrophic lateral sclerosis. Arch. Neurol. 68, 739–742 (2011).
Van Damme, P. et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology 76, 2066–2072 (2011).
Ross, O. A. et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum. Mol. Genet. 20, 3207–3212 (2011).
Deng, H. X. et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature http://dx.doi.org/10.1038/nature10353.
Gunnarsson, L.-G., Dahlbom, K. & Strandman, E. Motor neuron disease and dementia reported among 13 members of a single family. Acta Neurol. Scand. 84, 429–433 (1991).
Morita, M. et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology 66, 839–844 (2006).
Vance, C. et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2–213. Brain 129, 868–876 (2006).
Pearson, J. P. et al. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J. Neurol. 258, 647–655 (2011).
Laaksovirta, H. et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome wide association study. Lancet Neurol. 9, 978–985 (2010).
Boxer, A. L. et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD–ALS family. J. Neurol. Neurosurg. Psychiatry 82, 196–203 (2011).
Rollinson, S. et al. Frontotemporal lobar degeneration genome wide association study replication confirms a risk locus shared with amyotrophic lateral sclerosis. Neurobiol. Aging. 32, 758.e1–758.e7 (2011).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron http://dx.doi.org/10.1016/j.neuron.2011.09.011.
Renton, A. E. et al. A hexonucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS–FTD. Neuron http://dx.doi.org/10.1016/j.neuron.2011.09.010.
Al-Chalabi, A. et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J. Neurol. Neurosurg. Psychiatry 81, 1324–1326 (2010).
Haverkamp, L. J., Appel, V. & Appel, S. H. Natural history of amyotrophic lateral sclerosis in a database population. Validation of a scoring system and a model for survival prediction. Brain 118, 707–719 (1995).
Czaplinski, A., Steck, A. J., Andersen, P. M. & Weber, M. Flail arm syndrome: a clinical variant of amyotrophic lateral sclerosis. Eur. J. Neurol. 11, 567–568 (2004).
Riggs, J. E. Longitudinal Gompertzian analysis of amyotrophic lateral sclerosis mortality in the US, 1977–1986: evidence for an inherently susceptible subset. Mech. Ageing Dev. 55, 207–220 (1990).
Hanby, M. F. et al. The risk to relatives of patients with sporadic amyotrophic lateral sclerosis. Brain http://dx.doi.org/10.1093/brain/AWR248.
Falconer, D. S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 29, 51–76 (1965).
Edwards, J. H. The simulation of mendelism. Acta Genet. Stat. Med. 10, 63–70 (1960).
Fang, F. et al. Familial aggregation of amyotrophic lateral sclerosis. Ann. Neurol. 66, 94–99 (2009).
van Es, M. A. et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 41, 1083–1087 (2009).
Shatunov, A. et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 9, 986–994 (2010).
Van Landeghem, G. F. et al. Manganese-containing superoxide dismutase signal sequence polymorphism associated with sporadic motor neuron disease. Eur. J. Neurol. 6, 639–644 (1999).
Robberecht, W. et al. D90A heterozygosity for the SOD1 gene is associated with familial and apparently sporadic amyotrophic lateral sclerosis. Neurology 47, 1336–1339 (1996).
Battistini, S. et al. SOD1 mutations in amyotrophic lateral sclerosis. Results from a multicenter Italian study. J. Neurol. 252, 782–788 (2005).
Masé, G. et al. ALS with variable phenotypes in a six-generation family caused by leu144phe mutation in the SOD1 gene. J. Neurol. Sci. 191, 11–18 (2001).
Turner, M. R. et al. Distinct cerebral lesions in sporadic and 'D90A' SOD1 ALS: studies with [11C]flumazenil PET. Brain 128, 1323–1329 (2005).
Boukaftane, Y. et al. Identification of six novel SOD1 gene mutations in familial amyotrophic lateral sclerosis. Can. J. Neurol. Sci. 25, 192–196 (1998).
Hayward, C., Brock, D. J., Minns, R. A. & Swingler, R. J. Homozygosity for Asn86Ser mutation in the CuZn-superoxide dismutase gene produces a severe clinical phenotype in a juvenile onset case of familial amyotrophic lateral sclerosis. J. Med. Genet. 35, 174 (1998).
Kato, M. et al. Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci. Lett. 312, 165–168 (2001).
Parkinson, N. et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67, 1074–1077 (2006).
Schymick, J. et al. Progranulin mutations and ALS or ALS–FTD phenotypes. J. Neurol. Neurosurg. Psychiatry 78, 754–756 (2007).
Chow, C. Y. et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am. J. Hum. Genet. 84, 85–88 (2009).
Luty, A. A. et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration–motor neuron disease. Ann. Neurol. 68, 639–649 (2010).
Johnson, J. O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010).
Phukan, J. et al. Huntington's disease presenting as amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 11, 405–407 (2010).
Praline, J. et al. CADASIL and ALS: a link? Amyotroph. Lateral Scler. 11, 399–401 (2010).
Parton, M. J. et al. D90A-SOD1 mediated amyotrophic lateral sclerosis: a single founder for all cases with evidence for a Cis-acting disease modifier in the recessive haplotype. Hum. Mutat. 20, 473 (2002).
Broom, W. J. et al. SOD1A4V-mediated ALS: absence of a closely linked modifier gene and origination in Asia. Neurosci. Lett. 430, 241–245 (2008).
Saeed, M. et al. Age and founder effect of SOD1 A4V mutation causing ALS. Neurology 72, 1634–1639 (2009).
Rabe, M. et al. The epidemiology of CuZn-SOD mutations in Germany: a study of 217 families. J. Neurol. 257, 1298–1302 (2010).
Niemann, S. et al. Familial ALS in Germany: origin of the R115G SOD1 mutation by a founder effect. J. Neurol. Neurosurg. Psychiatry 75, 1186–1188 (2004).
Andersen, P. M. et al. EFNS task force on management of amyotrophic lateral sclerosis: guidelines for diagnosing and clinical care of patients and relatives. Eur. J. Neurol. 12, 921–938 (2005).
Safety, Tolerability, and Activity Study of ISIS SOD1Rx to Treat Familial Amyotrophic Lateral Sclerosis (ALS) Caused by SOD1 Gene Mutations (SOD-1). ClinicalTrials.gov [online], (2011).
Hand, C. K. et al. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am. J. Hum. Genet. 70, 251–256 (2002).
Orlacchio, A. et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain 133, 591–598 (2010).
Sapp, P. C. et al. Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am. J. Hum. Genet. 73, 397–403 (2003).
International HapMap Project [online], (2011).
Münch, C. et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology 63, 724–726 (2004).
van Es, M. A. et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 40, 29–31 (2008).
Cronin, S. et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet. 17, 768–774 (2008).
Ticozzi, N. et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156, 285–290 (2011).
Lambrechts, D. et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat. Genet. 34, 383–394 (2003).
Figlewicz, D. A. et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum. Mol. Genet. 3, 1757–1761 (1994).
Tomkins, J. et al. Novel insertion in the KSP region of the neurofilament heavy gene in amyotrophic lateral sclerosis (ALS). Neuroreport 9, 3967–3970 (1998).
Al-Chalabi, A. et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum. Mol. Genet. 8, 157–164 (1999).
Vechio, J. D., Bruijn, L. I., Xu, Z., Brown, R. H. Jr & Cleveland, D. Sequence variants in human neurofilament proteins: absence of linkage to familial amyotrophic lateral sclerosis. Ann. Neurol. 40, 603–610 (1996).
Rooke, K., Figlewicz, D. A., Han, F. Y. & Rouleau, G. A. Analysis of the KSP repeat of the neurofilament heavy subunit in familial amyotrophic lateral sclerosis. Neurology 46, 789–790 (1996).
Corcia, P. et al. Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann. Neurol. 51, 243–246 (2002).
Veldink, J. H. et al. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology 65, 820–825 (2005).
Simpson, C. L. et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481 (2009).
Slowik, A. et al. Paraoxonase gene polymorphisms and sporadic ALS. Neurology 67, 766–770 (2006).
Wang, X. S. et al. Increased incidence of the Hfe mutation in amyotrophic lateral sclerosis and related cellular consequences. J. Neurol. Sci. 227, 27–33 (2004).
Yen, A. A., Simpson, E. P., Henkel, J. S., Beers, D. R. & Appel, S. H. HFE mutations are not strongly associated with sporadic ALS. Neurology 62, 1611–1612 (2004).
Landers, J. E. et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA 106, 9004–9009 (2009).
Zetterberg, H., Jacobsson, J., Rosengren, L., Blennow, K. & Andersen, P. M. Association of APOE with age at onset of sporadic amyotrophic lateral sclerosis. J. Neurol. Sci. 273, 67–69 (2008).
Li, Y. J. et al. Apolipoprotein E is associated with age at onset of amyotrophic lateral sclerosis. Neurogenetics 5, 209–213 (2004).
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, made substantial contributions to discussions of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary table 1
Frequency of FALS in epidemiological studies (PDF 115 kb)
Supplementary Table 2
Frequency of SOD1 mutations in ALS (PDF 106 kb)
Supplementary Box 1
Guidelines for presymptomatic genetic testing in amyotrophic lateral sclerosis (PDF 88 kb)
Rights and permissions
About this article
Cite this article
Andersen, P., Al-Chalabi, A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know?. Nat Rev Neurol 7, 603–615 (2011). https://doi.org/10.1038/nrneurol.2011.150
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneurol.2011.150
This article is cited by
-
Cerebrospinal fluid and blood exosomes as biomarkers for amyotrophic lateral sclerosis; a systematic review
Diagnostic Pathology (2024)
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
Prospects for gene replacement therapies in amyotrophic lateral sclerosis
Nature Reviews Neurology (2023)
-
Predictors for progression in amyotrophic lateral sclerosis associated to SOD1 mutation: insight from two population-based registries
Journal of Neurology (2023)
-
Evaluating the causal association between microRNAs and amyotrophic lateral sclerosis
Neurological Sciences (2023)