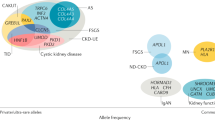
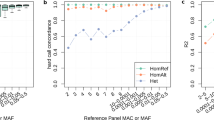
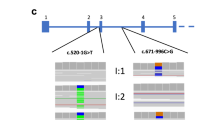
Key Points
-
Next-generation sequencing has enabled increasingly accurate and cost-effective methods for mutation detection in patients with inherited kidney disorders
-
As renal ciliopathies and congenital anomalies of the kidney and urinary tract are highly heterogeneous disorders, high-throughput sequencing approaches are required for identification of causal genes in research and diagnostics
-
Challenges for implementation of next-generation sequencing in clinical practice include ethical considerations and accurate interpretation of genetic variants
-
National and international multidisciplinary collaborations involving nephrologists, geneticists, biologists and bioinformaticians are crucial to enable elucidation of the genetic backgrounds of inherited kidney diseases in the research and diagnostics settings
Abstract
The advent of next-generation sequencing technologies has enabled genetic nephrology research to move beyond single gene analysis to the simultaneous investigation of hundreds of genes and entire pathways. These new sequencing approaches have been used to identify and characterize causal factors that underlie inherited heterogeneous kidney diseases such as nephronophthisis and congenital anomalies of the kidney and urinary tract. In this Review, we describe the development of next-generation sequencing in basic and clinical research and discuss the implementation of this novel technology in routine patient management. Widespread use of targeted and nontargeted approaches for gene identification in clinical practice will require consistent phenotyping, appropriate disease modelling and collaborative efforts to combine and integrate data analyses. Next-generation sequencing is an exceptionally promising technique that has the potential to improve the management of patients with inherited kidney diseases. However, identifying the molecular mechanisms that lead to renal developmental disorders and ciliopathies is difficult. A major challenge in the near future will be how best to integrate data obtained using next-generation sequencing with personalized medicine, including use of high-throughput disease modelling as a tool to support the clinical diagnosis of kidney diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mong Hiep, T. T. et al. Clinical characteristics and outcomes of children with stage 3–5 chronic kidney disease. Pediatr. Nephrol. 25, 935–940 (2010).
Greenbaum, L. A., Warady, B. A. & Furth, S. L. Current advances in chronic kidney disease in children: growth, cardiovascular, and neurocognitive risk factors. Semin. Nephrol. 29, 425–434 (2009).
Hildebrandt, F. Genetic kidney diseases. Lancet 375, 1287–1295 (2010).
Neveling, K. et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum. Mutat. 34, 1721–1726 (2013).
Pei, Y. & Watnick, T. Diagnosis and screening of autosomal dominant polycystic kidney disease. Adv. Chronic Kidney Dis. 17, 140–152 (2010).
Tory, K. et al. High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J. Am. Soc. Nephrol. 18, 1566–1575 (2007).
Hoefele, J. et al. Evidence of oligogenic inheritance in nephronophthisis. J. Am. Soc. Nephrol. 18, 2789–2795 (2007).
Sanger, F. & Coulson, A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–448 (1975).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977).
Thomas, R. et al. Identification of mutations in the repeated part of the autosomal dominant polycystic kidney disease type 1 gene, PKD1, by long-range PCR. Am. J. Hum. Genet. 65, 39–49 (1999).
Bamshad, M. J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 (2011).
Hehir-Kwa, J. Y., Pfundt, R., Veltman, J. A. & de Leeuw, N. Pathogenic or not? Assessing the clinical relevance of copy number variants. Clin. Genet. 84, 415–421 (2013).
Drmanac, R. The advent of personal genome sequencing. Genet. Med. 13, 188–190 (2011).
Need, A. C. et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J. Med. Genet. 49, 353–361 (2012).
Yang, Y. et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369, 1502–1511 (2013).
Gilissen, C., Hoischen, A., Brunner, H. G. & Veltman, J. A. Unlocking Mendelian disease using exome sequencing. Genome Biol. 12, 228 (2011).
Otto, E. A. et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 42, 840–850 (2010).
Lovric, S. et al. Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin. J. Am. Soc. Nephrol. http://dx.doi.org/10.2215/CJN.09010813.
Nijman, I. J. et al. Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat. Methods 7, 913–915 (2010).
Cummings, N. et al. Combining target enrichment with barcode multiplexing for high throughput SNP discovery. BMC Genomics 11, 641 (2010).
Harakalova, M. et al. Multiplexed array-based and in-solution genomic enrichment for flexible and cost-effective targeted next-generation sequencing. Nat. Protoc. 6, 1870–1886 (2011).
Teer, J. K. et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 20, 1420–1431 (2010).
Turner, E. H., Ng, S. B., Nickerson, D. A. & Shendure, J. Methods for genomic partitioning. Annu. Rev. Genomics Hum. Genet. 10, 263–284 (2009).
Frank, M., Prenzler, A., Eils, R. & Graf von der Schulenburg, J. M. Genome sequencing: a systematic review of health economic evidence. Health Econ. Rev. 3, 29 (2013).
Fuchs, S. A. et al. Application of exome sequencing in the search for genetic causes of rare disorders of copper metabolism. Metallomics 4, 606–613 (2012).
Gilissen, C., Hoischen, A., Brunner, H. G. & Veltman, J. A. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 20, 490–497 (2012).
Zhang, X. Exome sequencing greatly expedites the progressive research of Mendelian diseases. Front. Med. 8, 42–57 (2014).
International HapMap 3 Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 1, 40–47 (2000).
Jouan, L., Gauthier, J., Dion, P. A. & Rouleau, G. A. Rare variants in complex traits: novel identification strategies and the role of de novo mutations. Hum. Hered. 74, 215–225 (2012).
Veltman, J. A. & Brunner, H. G. De novo mutations in human genetic disease. Nat. Rev. Genet. 13, 565–575 (2012).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Vissers, L. E. et al. A de novo paradigm for mental retardation. Nat. Genet. 42, 1109–1112 (2010).
Xu, B. et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 44, 1365–1369 (2012).
Petrovski, S., Wang, Q., Heinzen, E. L., Allen, A. S. & Goldstein, D. B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709 (2013).
NHLBI Exome Sequencing Project. Exome Variant Server. evs.gs.washington.edu [online], (2014).
Dorschner, M. O. et al. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am. J. Hum. Genet. 93, 631–640 (2013).
Abdul-Karim, R. et al. Disclosure of incidental findings from next-generation sequencing in pediatric genomic research. Pediatrics 131, 564–571 (2013).
Christenhusz, G. M., Devriendt, K. & Dierickx, K. To tell or not to tell? A systematic review of ethical reflections on incidental findings arising in genetics contexts. Eur. J. Hum. Genet. 21, 248–255 (2013).
Lolkema, M. P. et al. Ethical, legal, and counseling challenges surrounding the return of genetic results in oncology. J. Clin. Oncol. 31, 1842–1848 (2013).
Bredenoord, A. L., Kroes, H. Y., Cuppen, E., Parker, M. & van Delden, J. J. Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet. 27, 41–47 (2011).
McCarthy, H. J. et al. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin. J. Am. Soc. Nephrol. 8, 637–648 (2013).
Tan, A. Y. et al. molecular diagnosis of autosomal dominant polycystic kidney disease using next-generation sequencing. J. Mol. Diagn. 16, 216–228 (2013).
Below, J. E. et al. Whole-genome analysis reveals that mutations in inositol polyphosphate phosphatase-like 1 cause opsismodysplasia. Am. J. Hum. Genet. 92, 137–143 (2013).
Hildebrandt, F. et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 5, e1000353 (2009).
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA 106, 19096–19101 (2009).
Katsanis, S. H. & Katsanis, N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 14, 415–426 (2013).
Hildebrandt, F., Benzing, T. & Katsanis, N. Ciliopathies. N. Engl. J. Med. 364, 1533–1543 (2011).
Eggenschwiler, J. T. & Anderson, K. V. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345–373 (2007).
Barr, M. M. & Sternberg, P. W. A polycystic kidney-disease gene homologue required for male mating behaviour in, C. elegans. Nature 401, 386–389 (1999).
The European Polycystic Kidney Disease Consortium. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77, 881–894 (1994).
Gunay-Aygun, M. Liver and kidney disease in ciliopathies. Am. J. Med. Genet. C Semin. Med. Genet. 151C, 296–306 (2009).
Hurd, T. W. & Hildebrandt, F. Mechanisms of nephronophthisis and related ciliopathies. Nephron Exp. Nephrol. 118, e9–e14 (2011).
Hildebrandt, F., Attanasio, M. & Otto, E. Nephronophthisis: disease mechanisms of a ciliopathy. J. Am. Soc. Nephrol. 20, 23–35 (2009).
Gagnadoux, M. F., Bacri, J. L., Broyer, M. & Habib, R. Infantile chronic tubulo-interstitial nephritis with cortical microcysts: variant of nephronophthisis or new disease entity? Pediatr. Nephrol. 3, 50–55 (1989).
Blowey, D. L., Querfeld, U., Geary, D., Warady, B. A. & Alon, U. Ultrasound findings in juvenile nephronophthisis. Pediatr. Nephrol. 10, 22–24 (1996).
Chung, E. M., Conran, R. M., Schroeder, J. W., Rohena-Quinquilla, I. R. & Rooks, V. J. From the radiologic pathology archives: pediatric polycystic kidney disease and other ciliopathies: radiologic-pathologic correlation. Radiographics 34, 155–178 (2014).
Waldherr, R., Lennert, T., Weber, H. P., Fodisch, H. J. & Scharer, K. The nephronophthisis complex. A clinicopathologic study in children. Virchows Arch. A Pathol. Anat. Histol. 394, 235–254 (1982).
Soliman, N. A. et al. Clinical characterization and NPHP1 mutations in nephronophthisis and associated ciliopathies: a single center experience. Saudi J. Kidney Dis. Transpl. 23, 1090–1098 (2012).
Hoff, S. et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet. 45, 951–956 (2013).
Simms, R. J., Hynes, A. M., Eley, L. & Sayer, J. A. Nephronophthisis: a genetically diverse ciliopathy. Int. J. Nephrol. 2011, 527137 (2011).
Arts, H. H. & Knoers, N. V. Current insights into renal ciliopathies: what can genetics teach us? Pediatr. Nephrol. 28, 863–874 (2013).
Attanasio, M. et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 39, 1018–1024 (2007).
Halbritter, J. et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J. Med. Genet. 49, 756–767 (2012).
Halbritter, J. et al. Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum. Genet. 132, 865–884 (2013).
Otto, E. A. et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J. Med. Genet. 48, 105–116 (2011).
Yuan, S. & Sun, Z. Expanding horizons: ciliary proteins reach beyond cilia. Annu. Rev. Genet. 47, 353–376 (2013).
Gee, H. Y. et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int. (2013).
Bredrup, C. et al. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am. J. Hum. Genet. 89, 634–643 (2011).
Chaki, M. et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150, 533–548 (2012).
Lee, J. H. et al. Evolutionarily assembled cis-regulatory module at a human ciliopathy locus. Science 335, 966–969 (2012).
Edvardson, S. et al. Joubert syndrome 2 (JBTS2) in Ashkenazi Jews is associated with a TMEM216 mutation. Am. J. Hum. Genet. 86, 93–97 (2010).
Nasonkin, I. O. et al. Conditional knockdown of DNA methyltransferase 1 reveals a key role of retinal pigment epithelium integrity in photoreceptor outer segment morphogenesis. Development 140, 1330–1341 (2013).
Nishiguchi, K. M. et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc. Natl Acad. Sci. USA 110, 16139–16144 (2013).
Loane, M. et al. Paper 4: EUROCAT statistical monitoring: identification and investigation of ten year trends of congenital anomalies in Europe. Birth Defects Res. A Clin. Mol. Teratol. 91 (Suppl. 1), S31–S43 (2011).
Melo, B. F. et al. Early risk factors for neonatal mortality in CAKUT: analysis of 524 affected newborns. Pediatr. Nephrol. 27, 965–972 (2012).
Groothoff, J. W. et al. Social consequences in adult life of end-stage renal disease in childhood. J. Pediatr. 146, 512–517 (2005).
European Renal Association–European dialysis and Transplant Association. ERA–EDTA Registry Annual Report 2011. era-edta-reg.org [online], (2011).
Madariaga, L. et al. Severe prenatal renal anomalies associated with mutations in HNF1B or PAX2 genes. Clin. J. Am. Soc. Nephrol. 8, 1179–1187 (2013).
Belk, R. A. et al. A family study and the natural history of prenatally detected unilateral multicystic dysplastic kidney. J. Urol. 167, 666–669 (2002).
Bulum, B. et al. High frequency of kidney and urinary tract anomalies in asymptomatic first-degree relatives of patients with CAKUT. Pediatr. Nephrol. 28, 2143–2147 (2013).
Horikawa, Y. et al. Mutation in hepatocyte nuclear factor-1β gene (TCF2) associated with MODY. Nat. Genet. 17, 384–385 (1997).
Winyard, P. & Chitty, L. S. Dysplastic kidneys. Semin. Fetal Neonatal Med. 13, 142–151 (2008).
Little, M., Georgas, K., Pennisi, D. & Wilkinson, L. Kidney development: two tales of tubulogenesis. Curr. Top. Dev. Biol. 90, 193–229 (2010).
Schedl, A. Renal abnormalities and their developmental origin. Nat. Rev. Genet. 8, 791–802 (2007).
Hwang, D. Y. et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. http://dx.doi.org/10.1038/ki.2013.508.
Humbert, C. et al. Integrin α8 recessive mutations are responsible for bilateral renal agenesis in humans. Am. J. Hum. Genet. 94, 288–294 (2014).
Naseri, M., Ghiggeri, G. M., Caridi, G. & Abbaszadegan, M. R. Five cases of severe vesico-ureteric reflux in a family with an X-linked compatible trait. Pediatr. Nephrol. 25, 349–352 (2010).
Kerecuk, L. et al. Autosomal dominant inheritance of non-syndromic renal hypoplasia and dysplasia: dramatic variation in clinical severity in a single kindred. Nephrol. Dial. Transplant. 22, 259–263 (2007).
Fletcher, J., McDonald, S., Alexander, S. I. & Australian & New Zealand Pediatric Nephrology Association. Prevalence of genetic renal disease in children. Pediatr. Nephrol. 28, 251–256 (2013).
Renkema, K. Y. et al. Novel perspectives for investigating congenital anomalies of the kidney and urinary tract (CAKUT). Nephrol. Dial. Transplant. 26, 3843–3851 (2011).
Sanna-Cherchi, S. et al. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet. 91, 987–997 (2012).
Gbadegesin, R. A. et al. TNXB mutations can cause vesicoureteral reflux. J. Am. Soc. Nephrol. 24, 1313–1322 (2013).
Saisawat, P. et al. Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int. http://dx.doi.org/10.1038/ki.2013.417.
Sanna-Cherchi, S. et al. Mutations in DSTYK and dominant urinary tract malformations. N. Engl. J. Med. 369, 621–629 (2013).
Vivante, A. et al. Renal hypodysplasia associates with a WNT4 variant that causes aberrant canonical WNT signaling. J. Am. Soc. Nephrol. 24, 550–558 (2013).
Djuric, T. et al. MMP-1 and -3 haplotype is associated with congenital anomalies of the kidney and urinary tract. Pediatr. Nephrol. 29, 879–884 (2014).
Barak, H. et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell 22, 1191–1207 (2012).
Kaku, Y. et al. Islet1 deletion causes kidney agenesis and hydroureter resembling CAKUT. J. Am. Soc. Nephrol. 24, 1242–1249 (2013).
Saisawat, P. et al. Identification of two novel CAKUT-causing genes by massively parallel exon resequencing of candidate genes in patients with unilateral renal agenesis. Kidney Int. 81, 196–200 (2012).
de Groot, T. et al. Lithium causes G2 arrest of renal principal cells. J. Am. Soc. Nephrol. 25, 501–510 (2014).
Sang, L. et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 (2011).
Choi, H. J. et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell 51, 423–439 (2013).
Luijten, M. N. et al. Birt-Hogg-Dube syndrome is a novel ciliopathy. Hum. Mol. Genet. 22, 4383–4397 (2013).
Ebarasi, L., Oddsson, A., Hultenby, K., Betsholtz, C. & Tryggvason, K. Zebrafish: a model system for the study of vertebrate renal development, function, and pathophysiology. Curr. Opin. Nephrol. Hypertens. 20, 416–424 (2011).
Kaufman, C. K., White, R. M. & Zon, L. Chemical genetic screening in the zebrafish embryo. Nat. Protoc. 4, 1422–1432 (2009).
Basten, S. G. et al. Mutations in LRRC50 predispose zebrafish and humans to seminomas. PLoS Genet. 9, e1003384 (2013).
Ross, A. J. et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat. Genet. 37, 1135–1140 (2005).
Zaghloul, N. A. & Katsanis, N. Zebrafish assays of ciliopathies. Methods Cell Biol. 105, 257–272 (2011).
Feeney, M. M. & Rosenblum, N. D. Urinary tract pacemaker cells: current knowledge and insights from nonrenal pacemaker cells provide a basis for future discovery. Pediatr. Nephrol. 4, 629–635 (2013).
Burn, S. F. et al. Calcium/NFAT signalling promotes early nephrogenesis. Dev. Biol. 352, 288–298 (2011).
Davies, J. A. et al. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum. Mol. Genet. 13, 235–246 (2004).
Barker, N. et al. Lgr5+ve stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2, 540–552 (2012).
Inoue, H., Nagata, N., Kurokawa, H. & Yamanaka, S. iPS cells: a game changer for future medicine. EMBO J. 33, 409–417 (2014).
Woltjen, K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009).
Meir, Y. J. et al. A versatile, highly efficient, and potentially safer piggyBac transposon system for mammalian genome manipulations. FASEB J. 27, 4429–4443 (2013).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Lam, A. Q. et al. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J. Am. Soc. Nephrol. http://dx.doi.org/10.1681/ASN.2013080831.
Takasato, M. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 16, 118–126 (2014).
Coene, K. L. et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum. Mol. Genet. 20, 3592–3605 (2011).
Boldt, K. et al. Disruption of intraflagellar protein transport in photoreceptor cilia causes Leber congenital amaurosis in humans and mice. J. Clin. Invest. 121, 2169–2180 (2011).
Mimura, I., Kanki, Y., Kodama, T. & Nangaku, M. Revolution of nephrology research by deep sequencing: ChIP-seq and RNA-seq. Kidney Int. 85, 31–38 (2014).
European project SYSCILIA. Syscilia [online], (2014).
van Dam, T. J. et al. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2, 7 (2013).
Ku, C. S. et al. Exome sequencing: dual role as a discovery and diagnostic tool. Ann. Neurol. 71, 5–14 (2012).
Robinson, P. N. et al. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610–615 (2008).
The Human Phenotype Ontology Project. Human Phenotype Ontology Website [online], (2014).
Gattone, V. H. 2nd, Wang, X., Harris, P. C. & Torres, V. E. Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat. Med. 9, 1323–1326 (2003).
Sayer, J. A. et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 38, 674–681 (2006).
Torres, V. E. et al. Effective treatment of an orthologous model of autosomal dominant polycystic kidney disease. Nat. Med. 10, 363–364 (2004).
Shillingford, J. M. et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl Acad. Sci. USA 103, 5466–5471 (2006).
Wang, S. & Dong, Z. Primary cilia and kidney injury: current research status and future perspectives. Am. J. Physiol. Renal Physiol. 305, F1085–F1098 (2013).
Tobin, J. L. & Beales, P. L. Restoration of renal function in zebrafish models of ciliopathies. Pediatr. Nephrol. 23, 2095–2099 (2008).
Cao, Y. et al. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc. Natl Acad. Sci. USA 106, 21819–21824 (2009).
Chamling, X. et al. Ectopic expression of human BBS4 can rescue Bardet-Biedl syndrome phenotypes in Bbs4 null mice. PLoS ONE 8, e59101 (2013).
McCampbell, K. K. & Wingert, R. A. New tides: using zebrafish to study renal regeneration. Transl. Res. 163, 109–122 (2014).
Genin, E., Feingold, J. & Clerget-Darpoux, F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum. Genet. 124, 357–368 (2008).
Iatropoulos, P. et al. Discordant phenotype in monozygotic twins with renal coloboma syndrome and a PAX2 mutation. Pediatr. Nephrol. 27, 1989–1993 (2012).
Zhang, Y. et al. BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum. Mol. Genet. 23, 40–51 (2014).
European Consortium for High-throughput Research in Rare Kidney Diseases. EURenOmics [online], (2014).
Bartlett, Y. K. & Coulson, N. S. An investigation into the empowerment effects of using online support groups and how this affects health professional/patient communication. Patient Educ. Couns. 83, 113–119 (2011).
Saunier, S. et al. A novel gene that encodes a protein with a putative src homology 3 domain is a candidate gene for familial juvenile nephronophthisis. Hum. Mol. Genet. 6, 2317–2323 (1997).
Hildebrandt, F. et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat. Genet. 17, 149–153 (1997).
Otto, E. A. et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 34, 413–420 (2003).
Olbrich, H. et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat. Genet. 34, 455–459 (2003).
Mollet, G. et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat. Genet. 32, 300–305 (2002).
Otto, E. et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am. J. Hum. Genet. 71, 1161–1167 (2002).
Otto, E. A. et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 37, 282–288 (2005).
Delous, M. et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39, 875–881 (2007).
Otto, E. A. et al. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J. Am. Soc. Nephrol. 19, 587–592 (2008).
Davis, E. E. et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 43, 189–196 (2011).
Halbritter, J. et al. Defects in the IFT-B component IFT172 cause Jeune and Mainzer-Saldino syndromes in humans. Am. J. Hum. Genet. 93, 915–925 (2013).
Acknowledgements
The authors' research was supported by grants from the European Community's Seventh Framework Programme FP7/2009 under grant agreements 305608 (EURenOmics) and 241955 (SYSCILIA) and the Consortium Programme of the Dutch Kidney Foundation under grant agreement CP11.18 (KOUNCIL).
Author information
Authors and Affiliations
Contributions
All authors researched the data, wrote the article, made a substantial contribution to discussions of the content and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Renkema, K., Stokman, M., Giles, R. et al. Next-generation sequencing for research and diagnostics in kidney disease. Nat Rev Nephrol 10, 433–444 (2014). https://doi.org/10.1038/nrneph.2014.95
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneph.2014.95