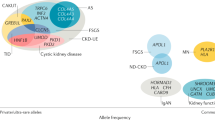
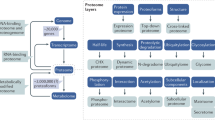
Abstract
Our understanding of the pathogenesis of most primary glomerular diseases, including IgA nephropathy, membranous nephropathy and focal segmental glomerulosclerosis, is limited. Advances in molecular technology now permit genome-wide, high-throughput characterization of genes and gene products from biological samples. Comprehensive examinations of the genome, transcriptome, proteome and metabolome (collectively known as omics analyses), have been applied to the study of IgA nephropathy, membranous nephropathy and focal segmental glomerulosclerosis in both animal models and human patients. However, most omics studies of primary glomerular diseases, with the exception of large genomic studies, have been limited by inadequate sample sizes and the lack of kidney-specific data sets derived from kidney biopsy samples. Collaborative efforts to develop a standardized approach for prospective recruitment of patients, scheduled monitoring of clinical outcomes, and protocols for sampling of kidney tissues will be instrumental in uncovering the mechanisms that drive these diseases. Integration of molecular data sets with the results of clinical and histopathological studies will ultimately enable these diseases to be characterized in a comprehensive and systematic manner, and is expected to improve the diagnosis and treatment of these diseases.
Key Points
-
Novel systems biology techniques that permit comprehensive molecular characterization of biological samples are improving our understanding of the molecular basis underlying primary glomerular diseases
-
Large-scale genomic studies of glomerular diseases have identified genetic loci that are associated with the development of these diseases and have enriched our understanding of their pathogenesis
-
Several novel monogenic forms of focal segmental glomerulosclerosis have been identified, which has improved the classification of hereditary and spontaneous variants of this disease and could potentially influence therapeutic decisions
-
Polymorphisms in APOL1 that increase the risk of developing focal segmental glomerulosclerosis and HIV-associated nephropathy have been identified in African American populations
-
Clinical application of the findings generated from systems-biology-based analysis is currently limited
-
Collaborative efforts for recruiting patients, monitoring clinical outcomes and developing standardized protocols for kidney tissue sampling, are needed to study rare primary glomerular diseases
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
United States Renal Data System. 2012 Atlas of CKD and ESRD [online], (2012).
Zuo, L. & Wang, M. Current burden and probable increasing incidence of ESRD in China. Clin. Nephrol. 74 (Suppl. 1), S20–S22 (2010).
Greenbaum, L. A., Benndorf, R. & Smoyer, W. E. Childhood nephrotic syndrome—current and future therapies. Nat. Rev. Nephrol. 8, 445–458 (2012).
He, J. C., Chuang, P. Y., Ma'ayan, A. & Iyengar, R. Systems biology of kidney diseases. Kidney Int. 81, 22–39 (2012).
Keller, B. J., Martini, S., Sedor, J. R. & Kretzler, M. A systems view of genetics in chronic kidney disease. Kidney Int. 81, 14–21 (2012).
Komorowsky, C. V., Brosius, F. C. 3rd, Pennathur, S. & Kretzler, M. Perspectives on systems biology applications in diabetic kidney disease. J. Cardiovasc. Transl. Res. 5, 491–508 (2012).
Jim, B., Santos, J., Spath, F. & Cijiang He, J. Biomarkers of diabetic nephropathy, the present and the future. Curr. Diabetes Rev. 8, 317–328 (2012).
Starkey, J. M. & Tilton, R. G. Proteomics and systems biology for understanding diabetic nephropathy. J. Cardiovasc. Transl. Res. 5, 479–490 (2012).
Hildebrandt, F. Genetic kidney diseases. Lancet 375, 1287–1295 (2010).
Heeringa, S. F. et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 121, 2013–2024 (2011).
Mele, C. et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N. Engl. J. Med. 365, 295–306 (2011).
Sanna-Cherchi, S. et al. Exome sequencing identified MYO1E and NEIL1 as candidate genes for human autosomal recessive steroid-resistant nephrotic syndrome. Kidney Int. 80, 389–396 (2011).
De Spiegelaere, W. et al. Quantitative mRNA expression analysis in kidney glomeruli using microdissection techniques. Histol. Histopathol. 26, 267–275 (2011).
Cohen, C. D., Frach, K., Schlondorff, D. & Kretzler, M. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 61, 133–140 (2002).
Gadegbeku, C. A. et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 83, 749–756 (2013).
Compendia Bioscience & Personalized Molecular Nephrology Research Laboratory. Nephromine [online], (2012).
Zurbig, P. et al. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes 61, 3304–3313 (2012).
Mullen, W., Delles, C. & Mischak, H. Urinary proteomics in the assessment of chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 20, 654–661 (2011).
Thongboonkerd, V. Biomarker discovery in glomerular diseases using urinary proteomics. Proteomics Clin. Appl. 2, 1413–1421 (2008).
Portilla, D., Schnackenberg, L. & Beger, R. D. Metabolomics as an extension of proteomic analysis: study of acute kidney injury. Semin. Nephrol. 27, 609–620 (2007).
Wishart, D. S. Metabolomics: a complementary tool in renal transplantation. Contrib. Nephrol. 160, 76–87 (2008).
Hirayama, A. et al. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal. Bioanal. Chem. 404, 3101–3109 (2012).
Donovan, M. J., Costa, J. & Cordon-Cardo, C. Systems pathology: a paradigm shift in the practice of diagnostic and predictive pathology. Cancer 115, 3078–3084 (2009).
Lai, K. N. Pathogenesis of IgA nephropathy. Nat. Rev. Nephrol. 8, 275–283 (2012).
Feehally, J. et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J. Am. Soc. Nephrol. 21, 1791–1797 (2010).
Gharavi, A. G. et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat. Genet. 43, 321–327 (2011).
Yu, X. Q. et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat. Genet. 44, 178–182 (2012).
Yang, C. et al. Genome-wide association study identifies TNFSF13 as a susceptibility gene for IgA in a South Chinese population in smokers. Immunogenetics 64, 747–753 (2012).
Cox, S. N. et al. Altered modulation of Wnt-β-catenin and PI3K/Akt pathways in IgA nephropathy. Kidney Int. 78, 396–407 (2010).
Preston, G. A. et al. Gene expression profiles of circulating leukocytes correlate with renal disease activity in IgA nephropathy. Kidney Int. 65, 420–430 (2004).
Woo, K. T. et al. Urotensin 2 and retinoic acid receptor α (RARA) gene expression in IgA nephropathy. Ann. Acad. Med. Singapore 39, 705–709 (2010).
Reich, H. N. et al. A molecular signature of proteinuria in glomerulonephritis. PLoS ONE 5, e13451 (2010).
Tokunaga, K. et al. Insulin-like growth factor binding protein-1 levels are increased in patients with IgA nephropathy. Biochem. Biophys. Res. Commun. 399, 144–149 (2010).
Flyvbjerg, A. Role of growth hormone, insulin-like growth factors (IGFs) and IGF-binding proteins in the renal complications of diabetes. Kidney Int. Suppl. 60, S12–S19 (1997).
Doublier, S. et al. Glomerulosclerosis in mice transgenic for human insulin-like growth factor-binding protein-1. Kidney Int. 57, 2299–2307 (2000).
Weber, J. A. et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 (2010).
Patnaik, S. K., Mallick, R. & Yendamuri, S. Detection of microRNAs in dried serum blots. Anal. Biochem. 407, 147–149 (2010).
Li, J. Y., Yong, T. Y., Michael, M. Z. & Gleadle, J. M. Review: the role of microRNAs in kidney disease. Nephrology (Carlton) 15, 599–608 (2010).
Kato, M., Arce, L. & Natarajan, R. MicroRNAs and their role in progressive kidney diseases. Clin. J. Am. Soc. Nephrol. 4, 1255–1266 (2009).
Wang, G. et al. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis. Markers 28, 79–86 (2010).
Wang, G. et al. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis. Markers 30, 171–179 (2011).
Serino, G., Sallustio, F., Cox, S. N., Pesce, F. & Schena, F. P. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J. Am. Soc. Nephrol. 23, 814–824 (2012).
Park, M. R. et al. Establishment of a 2D human urinary proteomic map in IgA nephropathy. Proteomics 6, 1066–1076 (2006).
Yokota, H. et al. Absence of increased α1-microglobulin in IgA nephropathy proteinuria. Mol. Cell Proteomics 6, 738–744 (2007).
Haubitz, M. et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 67, 2313–2320 (2005).
He, Q. et al. Urinary proteome analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry with magnetic beads for identifying the pathologic presentation of clinical early IgA nephropathy. J. Biomed. Nanotechnol. 8, 133–139 (2012).
Rocchetti, M. T. et al. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics 8, 206–216 (2008).
Maisonneuve, P. et al. Distribution of primary renal diseases leading to end-stage renal failure in the United States, Europe, and Australia/New Zealand: results from an international comparative study. Am. J. Kidney Dis. 35, 157–165 (2000).
Kerjaschki, D. & Farquhar, M. G. The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proc. Natl Acad. Sci. USA 79, 5557–5561 (1982).
Beck, L. H. Jr et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N. Engl. J. Med. 361, 11–21 (2009).
Stanescu, H. C. et al. Risk HLA-DQA1 and PLA2R1 alleles in idiopathic membranous nephropathy. N. Engl. J. Med. 364, 616–626 (2011).
Hauser, P. V. et al. Microarray and bioinformatics analysis of gene expression in experimental membranous nephropathy. Nephron Exp. Nephrol. 112, e43–e58 (2009).
Ngai, H. H. et al. Serial changes in urinary proteome profile of membranous nephropathy: implications for pathophysiology and biomarker discovery. J. Proteome Res. 5, 3038–3047 (2006).
Wang, L. et al. Autophagy can repair endoplasmic reticulum stress damage of the passive Heymann nephritis model as revealed by proteomics analysis. J. Proteomics 75, 3866–3876 (2012).
Beck, L. H. Jr et al. Rituximab-induced depletion of anti-PLA2R autoantibodies predicts response in membranous nephropathy. J. Am. Soc. Nephrol. 22, 1543–1550 (2011).
Qin, W. et al. Anti-phospholipase A2 receptor antibody in membranous nephropathy. J. Am. Soc. Nephrol. 22, 1137–1143 (2011).
Cravedi, P., Ruggenenti, P. & Remuzzi, G. Circulating anti-PLA2R autoantibodies to monitor immunological activity in membranous nephropathy. J. Am. Soc. Nephrol. 22, 1400–1402 (2011).
Gao, X. et al. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst. Biol. 6 (Suppl. 1), S14 (2012).
Kitiyakara, C., Kopp, J. B. & Eggers, P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin. Nephrol. 23, 172–182 (2003).
D'Agati, V. D., Kaskel, F. J. & Falk, R. J. Focal segmental glomerulosclerosis. N. Engl. J. Med. 365, 2398–2411 (2011).
Thomas, D. B. et al. Clinical and pathologic characteristics of focal segmental glomerulosclerosis pathologic variants. Kidney Int. 69, 920–926 (2006).
D'Agati, V. Pathologic classification of focal segmental glomerulosclerosis. Semin. Nephrol. 23, 117–134 (2003).
Lowik, M. M., Groenen, P. J., Levtchenko, E. N., Monnens, L. A. & van den Heuvel, L. P. Molecular genetic analysis of podocyte genes in focal segmental glomerulosclerosis—a review. Eur. J. Pediatr. 168, 1291–1304 (2009).
Brown, E. J. et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat. Genet. 42, 72–76 (2010).
Boyer, O. et al. INF2 mutations in Charcot–Marie–Tooth disease with glomerulopathy. N. Engl. J. Med. 365, 2377–2388 (2011).
Gharavi, A. G. et al. Mapping a locus for susceptibility to HIV-1-associated nephropathy to mouse chromosome 3. Proc. Natl Acad. Sci. USA 101, 2488–2493 (2004).
Kopp, J. B. et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 40, 1175–1184 (2008).
Kao, W. H. et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 40, 1185–1192 (2008).
Ghiggeri, G. M. et al. Genetics, clinical and pathological features of glomerulonephritis associated with mutations of nonmuscle myosin IIA (Fechtner syndrome). Am. J. Kidney Dis. 41, 95–104 (2003).
Sekine, T. et al. Patients with Epstein–Fechtner syndromes owing to MYH9 R702 mutations develop progressive proteinuric renal disease. Kidney Int. 78, 207–214 (2010).
Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010).
Kanji, Z. et al. Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J. Am. Soc. Nephrol. 22, 2091–2097 (2011).
Kopp, J. B. et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol. 22, 2129–2137 (2011).
Johnstone, D. B. et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS ONE 7, e51546 (2012).
Hodgin, J. B. et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am. J. Pathol. 177, 1674–1686 (2010).
Berger, S. I. & Iyengar, R. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 129–135 (2011).
Zhao, S. & Iyengar, R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu. Rev. Pharmacol. Toxicol. 52, 505–521 (2012).
Jin, Y. et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat. Med. 18, 580–588 (2012).
Lamb, J. et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Zhong, Y. et al. Renoprotective effect of combined inhibition of angiotensin-converting enzyme and histone deacetylase. J. Am. Soc. Nephrol. 24, 801–811 (2013).
Acknowledgements
S. Jiang, Z.-H. Liu and J. C. He are supported by Chinese 973 fund 2012CB517601NIH. J. C. He is supported by NIH grant numbers 1R01DK088541, 1R01DK078897, P01DK56492 and 1RC4DK090860, and a Veterans Administration Merit Award; P. Y. Chuang is supported by NIH grant number 5K08DK082760.
Author information
Authors and Affiliations
Contributions
S. Jiang, P. Y. Chuang, Z.-H. Liu and J. C. He contributed equally to writing the article. Z.-H. Liu and J. C. He also reviewed and edited the manuscript before submission. All authors contributed to researching data for the article and discussions of its content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Jiang, S., Chuang, P., Liu, ZH. et al. The primary glomerulonephritides: a systems biology approach. Nat Rev Nephrol 9, 500–512 (2013). https://doi.org/10.1038/nrneph.2013.129
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneph.2013.129
This article is cited by
-
Years of life lost and long-term outcomes due to glomerular disease in a Southeast Asian Cohort
Scientific Reports (2023)
-
Plasma acylcarnitines could predict prognosis and evaluate treatment of IgA nephropathy
Nutrition & Metabolism (2019)
-
Risk alleles for IgA nephropathy-associated SNPs conferred completely opposite effects to idiopathic membranous nephropathy in Chinese Han
Immunologic Research (2017)
-
Insights into kidney diseases from genome-wide association studies
Nature Reviews Nephrology (2016)
-
The etiology of glomerulonephritis: roles of infection and autoimmunity
Kidney International (2014)