Key Points
-
Interspecies transmission of influenza A viruses is the result of many factors. One of the key factors involved is a shift in the receptor-binding specificity of the virus, which is mostly determined by mutations in viral haemagglutinin (HA).
-
Recent structural studies have provided molecular insights into the HA–host receptor interactions that have enabled several influenza A virus subtypes to jump from avian to human hosts.
-
The combination of distinct amino acids at positions 225 and 190 of HA is important for determining the receptor-binding specificity of the H1 subtype.
-
The Q226L and G228S substitutions in the HA glycoproteins of the H2 and H3 subtypes are sufficient to change the binding preference from the avian receptor to the human receptor.
-
In an experimentally adapted H5 subtype, the Q226L substitution and loss of a glycosylation site near the receptor-binding site contribute to the shift in binding preference from the avian receptor to the human receptor.
-
In the H7 subtype, amino acid substitutions at positions 186 and 226 of HA increase the preference for binding to the human receptor.
Abstract
The recent emergence of the H7N9 avian influenza A virus and its ability to infect humans emphasize the epidemic and pandemic potential of these viruses. Interspecies transmission is the result of many factors, which ultimately lead to a change in the host tropism of the virus. One of the key factors involved is a shift in the receptor-binding specificity of the virus, which is mostly determined by mutations in the viral haemagglutinin (HA). In this Review, we discuss recent crystallographic studies that provide molecular insights into HA–host receptor interactions that have enabled several influenza A virus subtypes to 'jump' from avian to human hosts.
Similar content being viewed by others
Main
Influenza outbreaks have occurred since at least the Middle Ages, if not since ancient times1. In the past century, there were four severe influenza pandemics, in 1918 (Spanish flu), in 1957 (Asian flu), in 1968 (Hong Kong flu) and in 2009 (swine flu), as well as a moderate pandemic in 1977 (Russian flu)1. There is a high probability that we will face another influenza pandemic, but it is impossible to predict when it will happen, where it will originate, what virus subtype will cause the pandemic and the severity of such an outbreak. However, it is likely that new animal-derived influenza strains, particularly avian strains, will contribute to new pandemics2, and an increased understanding of the molecular mechanisms involved in determining influenza host tropism should facilitate such predictions in the future.
Influenza A virus is a zoonotic pathogen that can infect a broad range of species, including birds, pigs, dogs, horses, tigers and humans, causing annual epidemics (known as seasonal flu) and, at irregular intervals, pandemics (Box 1). The virus is an enveloped, single-stranded, negative-sense RNA virus with a segmented genome comprising eight gene segments3 that encode 16 proteins4,5 (Fig. 1a), although not all viruses express all 16 proteins. Haemagglutinin (HA) and neuraminidase (NA) are the two major viral envelope glycoproteins that recognize sialic acid (SA) on host cells. HA binds to sialylated host cell receptors and mediates membrane fusion, whereas NA removes sialyl residues from the membrane of infected cells and from viral membranes to enable budding and release of newly synthesized virus particles6. In the infected host, both HA and NA are targeted by neutralizing antibodies, and based on their antigenic properties, influenza type A viruses are classified into 18 HA subtypes (H1–H16 in wild waterfowl, and H17 and H18 in bats; note that the functions of the bat HA subtypes are currently unknown) and into 11 NA subtypes (N1–N9 in wild waterfowl, and N10 and N11 in bats)6,7,8,9,10.
a | Influenza A viruses are enveloped, single-stranded, negative-sense RNA viruses that contain eight gene segments that encode 16 proteins (although not all influenza viruses express all 16 proteins). The non-structural segment encodes the nuclear export protein NS2 and the host antiviral response antagonist NS1; the matrix segment encodes the matrix protein M1, the ion channel protein M2 and the M2-related protein M42 (which can functionally replace M2); the haemagglutinin (HA) segment encodes the receptor-binding glycoprotein HA; and the neuraminidase (NA) segment encodes NA (which cleaves sialic acid from cell surfaces). In addition, nucleoprotein (NP) and the components of the RNA-dependent RNA polymerase complex (PB1, PB2 and PA) are expressed from their respective genome segments. The two newly identified proteins N40 (the function of which is unknown93) and PA-X94, which represses cellular gene expression, are encoded by the PB1 and PA segments, respectively. Another two forms of PA (with amino-terminal truncations) have been found recently, named PA-N155 and PA-N182, which are likely to have important functions in the replication cycle of influenza A viruses5. In addition, some viruses express the pro-apoptotic protein PB1-F2, which is encoded by a second ORF in the PB1 segment. b | Virus infection is initiated by binding of the virus to sialylated host cell-surface receptors, and entry is mediated by endocytosis. In the host cell, fusion of viral and endosomal membranes occurs at low pH, which enables the release of the segmented viral genome into the cytoplasm. The viral genome is subsequently translocated to the nucleus, where it is transcribed and replicated. Following synthesis in the cytoplasm, viral proteins are assembled into viral ribonucleoproteins (vRNPs) in the nucleus. Export of vRNPs to the cytoplasm is mediated by M1 and NS2. Virus particles are assembled at the cell membrane, and the newly generated progeny virus buds into extracellular fluid.
To achieve interspecies transmission (known as a 'host jump'), influenza A virus must change its tropism to preferentially target new host species, and both viral and host factors have been implicated in this event11,12. The high mutation rate of the virus enables it to evolve rapidly and thereby overcome host barriers. All eight gene segments evolve continuously, but this evolution is most pronounced for the HA and NA glycoproteins. Evolution is achieved by two main mechanisms: genetic reassortment between different subtypes (known as antigenic shift if it occurs in either the HA or NA segments) and point mutations owing to antibody-mediated immune pressure (known as antigenic drift), including substitutions, deletions and insertions within the antibody-binding sites. This results in the generation of modified influenza virus genomes, which facilitates virus evasion of the host immune response.
HA proteins exhibit specific binding affinities for the different SA-linked glycoproteins that are expressed on cell-surface receptors. Avian viruses preferentially bind to SA linked to the terminal oligosaccharide by an α2,3 bond (which is referred to as the avian receptor), whereas human strains favour the α2,6-linked SA receptor (which is referred to as the human receptor). Specific amino acid mutations in HA lead to a change in receptor-binding preference and thus to altered host specificity and tropism. In addition to the structural determinants of viral HA and their corresponding receptors, other viral determinants contribute to host-specific adaptation, such as the balance between HA receptor-binding activity and NA-mediated release from infected cells, and amino acid substitutions in viral RNA polymerase (reviewed in Refs 11,12).
In this Review, we describe recent crystallographic studies that have identified the structural determinants of viral HA that enable interspecies transmission, and we also briefly consider corresponding changes in the host receptor. First, we provide a brief overview of the host cell receptors and viral HA proteins that are involved in the initial stages of influenza A virus infection. We then discuss recent crystallographic structures of the H1, H2, H3, H5 and H7 HA subtypes in complex with both avian and human receptors, with a focus on the amino acid substitutions in the receptor-binding site of HA that enable the host jump. Please note, all amino acid residues throughout the paper are numbered according to the H3 subtype (as is convention in the field), which enables the different virus subtypes to be compared.
The influenza virus life cycle
The virus initially binds to SA-linked host cell-surface receptors via the HA glycoprotein (see below) and, following entry into the cell by endocytosis13, the viral and endosomal membranes fuse under low pH conditions14,15,16 (Fig. 1b). The viral genome is subsequently released into the cytoplasm and migrates to the nucleus, where it is transcribed and replicated. Viral segments then associate with nucleoprotein (NP) to form viral ribonucleoproteins (vRNPs), which are exported to the cytoplasm for packaging with the assistance of matrix protein 1 (M1) and non-structural protein 2 (NS2; also known as NEP). NS1 is not included in the virion but is abundantly expressed in infected cells17. The vRNPs translocate to the cell membrane along with the envelope proteins, HA, NA, M1 and the ion channel protein M2, and virus particles are formed18. Finally, release of the virus particle from the host cell is mediated by NA, and the infection spreads to other host cells.
Host cell-surface receptors. The cell-surface receptors that influenza viruses bind to are glycolipids or glycoproteins that contain terminal SA moieties19,20,21. Glycoproteins contain two types of glycan modification, N-glycans and O-glycans12; N-linked glycan chains are attached to asparagine residues, whereas O-linked glycosylation occurs at serine or threoine residues. N-glycans, O-glycans and glycolipids have different core structures12; however, it is the distinct structural features of the terminal SA moieties attached to the glycan chains that are the key determinants of receptor-binding specificity, rather than differences in the core structures. The host receptors that influenza viruses bind to contain the three common terminal saccharides SA1, galactose (Gal2) and N-acetylglucosamine (GlcNAc3)22 (the numbers correspond to the position of the terminal saccharides), and the penultimate Gal is linked to either α2,3-SA or α2,6-SA. Previous studies have revealed that the entry of influenza viruses is reduced in the absence of sialylated N-glycans23,24, and the internalization of influenza viruses via macropinocytic endocytosis, but not uptake via clathrin-mediated endocytosis, is dependent on N-glycans24. Moreover, mouse cells that lack glycolipids can be efficiently infected with human H3N2 virus25, which suggests that glycolipids are not essential for virus binding. As most studies have been carried out in vitro with human viruses, the specific roles of sialylated N-glycans, O-glycans and glycolipids in vivo and their roles in the binding of avian viruses have not been established.
The α2,6-linked SA receptor is predominantly found in the upper respiratory tract (URT) in humans, and α2,3-linked SA receptors are expressed in the lower respiratory tract (LRT)26. Thus, human influenza virus replicates readily in the upper airway, whereas replication of the avian influenza virus mainly occurs in the LRT in humans, which explains why avian viruses occasionally infect humans.
HA glycoprotein. HA is the most abundant protein on the surface of the virion, and it mediates binding to the host receptor and fusion between the viral and host endosomal membranes. In addition, it is the primary target of neutralizing antibodies directed against the different viral subtypes. The HA precursor polypeptide (HA0) is a type 1 transmembrane glycoprotein of about 550 amino acids, with an amino-terminal signal sequence, a transmembrane domain near the carboxyl terminus and a short cytoplasmic tail. The HA0 precursor is activated by proteolytic cleavage into two disulphide-linked polypeptides, HA1 and HA2. Three monomers of HA1–HA2 form the mature 220 kDa homotrimeric HA protein.
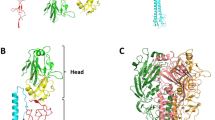
The X-ray crystal structure of the HA ectodomain was first reported in 1981 (Ref. 27). The HA trimer projects approximately 135 Å from the viral membrane and can be divided into two domains: the membrane-distal globular domain and the membrane-proximal stem domain (Fig. 2a). The receptor-binding site forms a shallow pocket at the tip of the globular domain and comprises three secondary structural elements and one base element6. The three secondary elements, namely the 130-loop, the 190-helix and the 220-loop (the numbers correspond to the amino acids in the H3 subtype), form the edges of the receptor-binding site, and four highly conserved residues (Y98, W153, H183 and Y195, of the H3 subtype) form the base. The SA moiety of the receptor typically forms several conserved hydrogen bonds with the 130-loop and the base residue Y98, and the remaining glycan moieties interact with the 220-loop or the 190-helix. In addition, the residues W153, H183 and Y195 contribute to receptor binding via van der Waals interactions.
a | The crystal structure of the ectodomain of haemagglutinin (HA) reveals two distinct domains: the globular domain and the stem domain. The receptor-binding site is located on the membrane-distal globular domain and forms a shallow pocket comprising three secondary elements: the 130-loop, 190-helix and 220-loop (the numbers correspond to the amino acids in the H3 subtype; see inset). Four highly conserved residues (Y98, W153, H183 and Y195) form the base element of the receptor-binding site (indicated in orange). The fusion peptide, which inserts into the host membrane during membrane fusion, is indicated. The structural figures were created using Protein Data Bank (PDB) accession 4JUG. b,c | The HA receptor analogues can be categorized into two types: the avian receptor analogue (α2,3-linked sialylated glycan receptor) (part b) and the human receptor analogue (α2,6-linked sialylated glycan receptor) (part c). Sialylated glycans of the host receptors that influenza viruses bind to contain the three terminal saccharides: the terminal sialic acid SA1, the galactose ring Gal2 (at position two relative to SA) and N-acetylglucosamine GlcNAc3 (at position three relative to SA). When bound by HA, the α2,3-linked SA receptor adopts a trans conformation and the hydrophilic glycosidic oxygen atom faces the 220-loop in the receptor-binding site (part b), whereas the α2,6-linked SA receptor adopts a cis conformation and the hydrophobic C6 atom points towards the 220-loop (part c). The structural figures are created using the PDB accessions 4JUH and 4JUJ.
The avian α2,3-linked SA receptor typically has an extended configuration in the HA-bound state, and the glycan rings interact with the 220-loop of HA, whereas the human α2,6-linked SA receptor displays a folded configuration, with the glycan rings interacting with the 190-helix (Fig. 2b,c). In most of the previously determined crystal structures of the HA–receptor complex, the α2,3-linked SA receptor adopts a trans conformation and the hydrophilic glycosidic oxygen atom faces the 220-loop, whereas the α2,6-linked SA receptor adopts a cis conformation and the hydrophobic C6 atom points towards the 220-loop. However, recent structural studies of the H7 subtype complexed with the avian receptor analogue challenge this dogma28. Both cis and trans conformations of the α2,3 SA moieties were observed in low-energy environments, which suggests that both conformations are stable in the HA-bound state28 (see below). The different conformations of α2,3-linked and α2,6-linked SA receptors require that the receptor-binding site of HA contains distinct amino acids: hydrophilic residues are required for HA binding to α2,3-linked SA, whereas hydrophobic residues are required for binding to α2,6-linked SA. Therefore, amino acid substitutions in the receptor-binding site of HA are required to facilitate the host jump. Only the H1, H2 and H3 subtypes have naturally adapted to infect humans. The specific amino acid substitutions that result in altered receptor-binding properties are distinct for each HA subtype (see below).
Receptor binding determinants of H1
Two severe flu pandemics were caused by the H1N1 subtype: the 1918 Spanish flu pandemic and the 2009 swine flu pandemic.
Crystal structures of H1–receptor complexes have been determined for HA proteins from avian, swine and human isolates29,30,31,32,33. Most of the avian H1 subtypes can bind to both α2,3- and α2,6-linked SA receptors, and the receptor-binding site in HA contains the residues E190 and G225 (Refs 32,33) (Fig. 3a). By contrast, in the H1 glycoproteins from two human-adapted isolates, the residues E190 and G225 are mutated to D190 and D225, and these HA glycoproteins specifically bind to the human α2,6-linked SA receptor (Fig. 3b). Binding to α2,6-linked SA receptors is mediated by hydrogen bonds between the glycan ring of the receptor and D190 and D225 in HA29,30. Crystal structures of a human receptor analogue in complex with HA showed that D190 interacts with GlcNAc3, whereas D225 contacts Gal2. Owing to differences in the overall configuration of the α2,6-linked SA receptor compared with the α2,3-linked SA receptor (folded versus extended), these interactions are absent in the crystal structure of the avian HA–receptor complex29,30.
Crystal structures of H1, H2 and H3 haemagglutinin (HA)–receptor complexes have revealed the structural basis for the switch in binding specificity. a | H1 HA from the avian subtype contains the residues E190 and G225 in the receptor-binding site, and can bind to both α2,3-linked (avian; shown in cyan) and α2,6-linked (human; shown in magenta) sialic acid (SA) receptors. The structural figures were created using Protein Data Bank (PDB) accessions 1RVX and 1RVZ. b | H1 HA proteins from two human isolates from the 1918 and 2009 pandemics contain the residues D190 and D225 and specifically bind to the human receptor. The crystal structure revealed that D190 interacts with N-acetylgalactosamine GlcNAc3, whereas D225 contacts the galactose ring Gal2.These interactions are absent in the avian HA–receptor complex owing to the different configurations of the human receptor (folded) compared with the avian receptor (extended)29,30. The structural figure was created using the PDB accession 4JTV. c | H1 HA mutants from later isolates from the 1918 and 2009 pandemics contain the residues D190 and G225 and can bind to both avian and human receptors. It is likely that G225 increases the flexibility of the 220-loop, which provides a suitable microenvironment to accommodate the extended configuration of the avian α2,3-linked SA receptor. The structural figures were created using PDB accessions 4JUH and 4JUJ. d | Avian HA proteins from the H2 and H3 subtypes contain the residues Q226 and G228, and can bind to both avian and human receptors. The structural figures were created using PDB accessions 2WR3 and 2WR4. e | HA proteins from human-adapted H2 and H3 subtypes contain the residues L226 and S228, and preferentially bind to the human receptor. L226 creates a hydrophobic environment that is favourable for the orientation of the hydrophobic C6 atom of the α2,6-linked SA receptor but is incompatible with the orientation of the hydrophilic glycosidic oxygen of the α2,3-linked SA receptor. The residue S228 forms a hydrogen bond with SA1, which increases the binding affinity of HA for the human receptor. Moreover, the avian H2 HA binds more efficiently to the human receptor than the avian H3 HA owing to hydrogen-bond interactions between N186 in H2 HA and Gal2, which are absent in H3 HA owing to the short side chain of S186. The structural figure was created using PDB accession 2WR7.
During the late stages of the 1918 and 2009 pandemics, D225G HA reversion mutants of both of these pandemic viruses emerged in patients34,35 (Fig. 3c). These D225G mutants (which retained the E190D mutation) had dual receptor-binding specificity, which was confirmed by surface plasmon resonance and glycan microarray experiments29,30 (Fig. 3c). Different structural explanations for the shift in receptor-binding specificity have been proposed. First, it has been suggested that the D225G substitution results in the loss of a salt bridge between D225 and K222 in HA, which relaxes the 220-loop and enables the Q226 residue to interact with the α2,3-linked SA receptor, which has not been observed in other H1 HA–receptor complexes30. Alternatively, it was proposed that the D225G substitution induces a distinct conformational change in the 220-loop as a result of an alteration in the backbone conformation of G225 (Ref. 29). Both models are plausible, and it is likely that both the loss of the salt bridge and the conformational change in G225 increase the flexibility of the 220-loop, which provides a suitable microenvironment to accommodate the extended configuration of the α2,3-linked SA receptor.
During adaptation of the H1 subtype to humans, other combinations of amino acid substitutions occurred. For example, HA from an early human H1 isolate (from 1934) was found to contain E190 and D225 (Refs 31,36), which enabled binding to both avian and human receptors, whereas HA from another isolate from the 2009 pandemic carried D190 and E225 and was specific for the human receptor30,34. Some of these intermediate substitutions are considered to be stepping stones on the pathways towards full specificity for the human receptor. The fact that earlier isolates have dual specificity and later pandemic isolates specifically bind to the human receptor highlights the remarkable capacity for these viruses to undergo a shift from dual specificity to human specificity.
In summary, the combination of distinct amino acids at positions 225 and 190 is important for determining the receptor-binding specificity of the H1 subtype. HA proteins that contain E190/G225, E190/D225 or D190/G225 in their receptor-binding site have dual receptor-binding specificity, whereas those that contain D190/D225 and D190/E225 preferentially bind to the human receptor. It is noteworthy that an amino acid substitution (alanine to serine) at position 138 was also found to be important for the host jump36; H1 isolates with dual-binding specificity have alanine at position 138, whereas those that are human receptor specific contain serine. This idea was proposed as early as 1989 (Ref. 36); however, structural evidence is still lacking.
Binding determinants of H2 and H3
The 1957 and 1968 pandemics were caused by influenza viruses of the H2N2 and H3N2 subtypes, respectively37. The human-adapted H2 and H3 HA glycoproteins both differ from their avian-specific HA counterparts in terms of two amino acid substitutions: Q226L and G228S (Refs 38,39). Avian H2 and H3 HA proteins can bind to both α2,3-linked and α2,6-linked SA receptors, whereas the human-adapted H2 and H3 HA proteins preferentially bind to the human receptor22,32,40 (Fig. 3d,e). Structural studies revealed that the residue L226 creates a hydrophobic environment that is incompatible with the orientation of the hydrophilic glycosidic oxygen of the α2,3-linked SA receptor but that is favourable for the orientation of the hydrophobic C6 atom of the α2,6-linked SA receptor22,32 (Fig. 2b,c). In addition, the residue S228 forms a hydrogen bond with SA1, which increases the binding affinity of HA for the human receptor22,32. The human receptor adopts a cis conformation when bound to both avian and human H2 and H3 HA proteins.
A recent paper explored the evolution of the receptor-binding properties of the HA glycoprotein from the H3N2 subtype41. Since its introduction into humans in 1968, the HA protein has continued to evolve. H3 viruses that were isolated after 1999 have reduced affinity for both human and avian receptors and, accordingly, these viruses display poor replicative potential in avian eggs and mammalian cell cultures42,43,44,45. Several key changes in the receptor-binding site of HA occurred41. Two sequential amino acid substitutions were found at position 225: between the years 2001 and 2002, the G225D substitution emerged, which was accompanied by the W222R substitution; and between the years 2004 and 2005, the D225N substitution was accompanied by the S193F substitution (while maintaining arginine at position 222). The residue at position 226 also underwent two mutations: the L226V substitution was detected before 2001, and in 2004, the V226I mutation emerged. Crystal structures of complexes formed between human receptor analogues and HA proteins (containing F193, R222, N225 and I226) from viruses isolated in 2004 and 2005 revealed substantial differences in the flexibility of the 220-loop compared with the crystal structure of the 1968 H3 HA in complex with the receptor. These conformational changes affect the interactions between the 220-loop and glycan moieties of the receptor analogue, which might explain the reduced affinity of these later strains for the human receptor41. The D225N substitution impairs the formation of hydrogen bonds between the side chain of residue D225 and Gal2, whereas the L226V and V226I mutations destabilize the interaction with the hydrophobic C6 atom in the glycosidic linkage of the human receptor, thus resulting in reduced affinity for the human receptor.
Electron density studies revealed that the avian H2 HA binds more efficiently to the human receptor than the avian H3 HA owing to hydrogen bond interactions between N186 in H2 HA and Gal2; these interactions are absent in H3 HA owing to the short side chain of S186 (Ref. 32). The N186 substitution might enable the H2 virus to gain an initial 'foothold' in the human host, and subsequent selective pressure could result in the Q226L and G228S substitutions, which might increase the binding affinity of the virus to the human receptor and reduce binding to the avian receptor.
Host jump of the H5 subtype
Although the highly pathogenic avian influenza (HPAI) virus H5N1 has caused several hundred sporadic cases of human infections since 1997 (Ref. 46), it has not yet acquired the ability to efficiently transmit between humans. There is currently no evidence to suggest that H5 has acquired the ability to preferentially bind the human receptor47,48,49, although recent studies have shown that it is possible to generate H5N1 mutants with a preference for the α2,6-linked SA receptor and the capacity for airborne transmission50,51,52. Two of these studies identified four artificially selected mutations in the HA protein that enable H5N1 viruses to break through the species barrier and to become transmissible by the airborne route in a ferret mammalian model system (which is an important experimental animal model for studying human influenza infections). In one study, H5N1 was genetically modified by site-directed mutagenesis and was then serially passaged in ferrets50, and in another study, a reassortant H5 HA–H1N1 virus was generated by combining a mutated HA from the H5N1 virus with the other seven gene segments from a 2009 pandemic H1N1 strain51 (the virus strains studied were A/Indonesia/5/2005 and A/Vietnam/1203/2004, respectively50,51). These experiments have provided some important clues about how H5 HA can efficiently bind to the human receptor, but they have engendered some controversy owing to the potential use of the mutated viruses as bioweapons or the threat they might pose if accidentally released. The amino acid substitutions that led to the host jump are N158D, N224K, Q226L and T318I in the strain from Vietnam, and H110Y, T160A, Q226L and G228S in the Indonesian strain. Thus, the only mutation that is shared by both is the Q226L substitution, which is located in the receptor binding site of HA50,51. Since then, the molecular basis of the receptor-binding specificity shift has been elucidated53,54,55. Crystal structures of the wild-type and mutant H5 HA proteins from A/Indonesia/5/2005, A/Vietnam/1194/2004 (in which the HA sequence is highly similar to A/Vietnam/1203/2004, with only one amino acid difference) and A/Vietnam/1203/2004 in complex with receptor analogues revealed that residue L226 creates a hydrophobic environment that is favourable for binding of the mutant HA proteins to the human receptor, but not to the avian receptor53,54,55,56,57 (Fig. 4a,b). This is similar to the observations for human-adapted H2 and H3 HA proteins (Fig. 3d,e). Further structural analysis indicated that binding of the HA mutant from the Indonesian strain to avian or human receptor analogues can lead to a trans to cis conformational switch53. In addition, the N158D or T160A substitutions in mutant H5 HA proteins from the strains from Vietnam or Indonesia, respectively, resulted in the loss of a glycosylation site near the receptor-binding site, which increased preference for the human receptor. Moreover, the T318I and H110Y substitutions from the Vietnam and Indonesia strains, respectively, increased HA thermostability, which is important for airborne transmissibility of virus among ferrets53,54,55. The T318I substitution does not affect receptor binding and instead stabilizes the position of the fusion peptide within the HA monomer54 (the fusion peptide consists of the hydrophobic HA2 residues 1–10, which are inserted into the target membrane during virus–membrane fusion (Fig. 1a)). By contrast, the H110Y substitution tightly connects the HA monomers via hydrogen bonding, which stabilizes the trimeric protein53. This indicates that several amino acid substitutions can lead to increased HA stability, which might facilitate airborne transmission.
Crystal structures of H5 and H7 haemagglutinin (HA) proteins in complex with avian and human receptor analogues. a | Wild-type H5 HA contains the residue Q226 and is glycosylated at residue N158. It has the capacity to bind preferentially to the avian receptor (α2,3-linked sialic acid (SA) receptor; shown in cyan) in a favourable trans conformation, but it binds weakly to the human receptor (α2,6-linked SA receptor; shown in magenta) in an unfavourable trans conformation. The hydrophilic residue Q226 provides a hydrophilic environment, which is compatible with the hydrophilic glycosidic oxygen atom of the avian receptor and incompatible with the hydrophobic C6 atom of the human receptor. Thus, the H5 HA preferentially binds the avian receptor. The structural figures were created using the Protein Data Bank (PDB) accessions 4K63 and 4K64. b | The ferret-transmissible mutant H5 HA (H5mut) has the residue L226 and residue 158 is deglycosylated. It binds preferentially to human receptor in a favourable cis conformation, whereas it binds weakly to the avian receptor in an unfavourable cis conformation. The hydrophobic residue L226 creates a hydrophobic environment, which is compatible with the hydrophobic C6 atom and incompatible with the hydrophilic glycosidic oxygen atom. Thus, H5mut preferentially binds to the human receptor. The structural figures were created using PDB accessions 4K66 and 4K67. c,d | The crystal structures of Anhui-H7N9 HA (AHH7, carrying the four amino acid substitutions S138A, G186V, T221P and Q226L) and an Anhui-H7N9 HA mutant (AHH7mut), in which L226 has been mutated to Q226, in complex with the avian or human receptor analogues provide insights into the structural basis of the shift in receptor binding. AHH7 and AHH7mut bind to both avian and human receptor analogues. The avian receptor analogues bind in different conformations to AHH7 (cis) and the AHH7mut carrying Q226 (trans), whereas the human receptor analogue binds to both proteins in a cis conformation. The four amino acid substitutions in AHH7 create a more hydrophobic environment that is favourable for binding to the human receptor. Furthermore, mutagenesis assays showed that the amino acid substitution Q226L is not solely responsible for the shift in receptor-binding preference of AHH7, and other substitutions also have a role. The structural figures were created using the PDB accessions 4KOM, 4KON, 4LKJ and 4LKK.
In addition, a recent study showed that a minimum set of five substitutions is sufficient to enable airborne transmission of the H5N1 virus in ferrets58. The five substitutions occurred in the RNA polymerase proteins (PB1 and PB2) and HA. The two substitutions in PB1 and PB2 together enhanced transcription and virus replication owing to increased polymerase activity58. These findings are consistent with a previous study in which the importance of RNA polymerase mutations in enabling the H5N1 host jump was also emphasized59. Moreover, the H110Y substitution in HA increased protein thermostability, and two further substitutions (T160A and Q226L (or G228S)) in HA changed the binding preference from avian to human receptors (Fig. 4a,b). Although HA proteins with single Q226L or G228S substitutions bound less efficiently to the human receptor compared with the double mutant, viruses with the single substitutions, which also contained the other substitutions in PB1,PB2, H110Y and T160A, were still transmissible58. These findings suggest that the ability of H5N1 to bind to the human receptor is not necessarily associated with its capacity to be transmitted via the airborne route.
Although the insights gained from studies in ferrets cannot be extrapolated for human-to-human transmission, the ferret transmission model is one of the best models for influenza research available today60. Some of the substitutions identified (such as E627K in PB2 and T160A in HA) are often found in natural virus isolates, and functionally equivalent substitutions exist for most of the identified substitutions in ferrets. These findings emphasize the risk of the emergence of transmissible H5N1 influenza A viruses and, as such, this needs to be monitored closely61.
Host jump of the H7 subtype
Since 1979, there have been sporadic cases of human infections with both low-pathogenic avian influenza (LPAI) and HPAI H7 viruses from Eurasian and North American lineages62,63,64,65,66,67,68,69,70,71. However, their inefficient transmissibility among humans has thus far prevented an epidemic or pandemic. Before 2013, the largest outbreak of an HPAI H7N7 virus occurred in 2003 in the Netherlands, with three cases of possible human-to-human virus transmission65.
In 2013, a novel avian H7N9 influenza virus that caused severe human infections was first identified in Shanghai and Anhui in China. This virus is an LPAI virus in domestic poultry, but it can cause severe respiratory diseases in humans72. The severity of the infection, preliminary epidemiology, environmental factors, virus origin and diversity have all been thoroughly investigated2,28,72,73,74,75. In addition, the pathogenicity and transmissibility of H7N9 have been evaluated in different animal models. Transmission experiments in the ferret model revealed that H7N9 viruses are efficiently spread between these animals via direct contact but are spread less efficiently by the airborne route76,77,78,79.
The HA proteins from avian H7N9 viruses preferentially bind to the avian receptor, but most have acquired the ability to also bind to human receptors28,80,81,82. The molecular basis underlying HA–receptor binding for the Anhui-H7N9 and Shanghai-H7N9 strains have been determined by crystallography28,82. In addition, similar studies were carried out to determine the structural basis for HA binding by comparing Anhui-H7N9 and one avian H7N3 virus isolate80. Anhui-H7N9 is the most prevalent strain in humans, whereas Shanghai-H7N9 was only isolated in a few cases83. Shanghai-H7N9 preferentially binds to the avian receptor analogue, whereas Anhui-H7N9 binds to both avian and human receptor analogues (although it has a higher affinity for the avian receptor)28,80 (Fig. 4c). This suggests that this change in receptor binding enabled Anhui-H7N9 to infect humans. However, glycan array analysis of the HA protein of A/Shanghai/2/2013 (the HA sequence is identical to Anhui-H7N9 HA) reveals negligible binding to human receptors and a strong preference for avian receptors81. This discrepancy may be due to the different assays used and should be further analysed in the future. A comparative analysis of Shanghai-H7N9 and Anhui-H7N9 HA proteins revealed that Anhui-H7N9 HA had evolved a total of eight amino acid substitutions, and structural analysis showed that four of these substitutions (S138A, G186V, T221P and Q226L) are located in the receptor-binding site28 (Fig. 4c). Interestingly, mutagenesis studies showed that the amino acid substitution Q226L is not solely responsible for the shift in receptor binding preference of Anhui-H7N9 HA and that the other amino acid substitutions in the receptor-binding site are likely to contribute. This is in contrast to H5N1, in which the Q226L substitution and the loss of a glycosylation site enabled binding to the human receptor. It is possible that Anhui-H7N9 HA acquired the ability to bind to the human receptor owing to the introduction of two bulky hydrophobic residues by the Q226L and G186V substitutions28,80. The crystal structures of Shanghai-H7N9 HA, Anhui-H7N9 HA and an Anhui-H7N9 HA mutant carrying the L226Q mutation, in their free forms and in complex with the avian or human receptor analogues, uncovered the structural basis of the shift in receptor binding28 (Fig. 4c,d). The four substitutions in Anhui-H7N9 HA (S138A, G186V, T221P and Q226L) create a hydrophobic region in the receptor-binding site, which enhances binding to the human receptor. For the first time, two conformations of the avian receptor analogue were observed: the avian receptor adopted a cis conformation in complex with Shanghai-H7N9 HA and a trans conformation when bound to the mutant Anhui-H7N9, despite the fact that both viruses carry the hydrophilic amino acid Q226. It has been reported that the avian receptor analogue can exist in two distinct conformations in aqueous solution, whereas the human receptor analogue mainly exists in the cis conformation84,85,86,87,88,89. These findings suggest that not only amino acid substitutions in HA but also the differential conformations of the human and avian receptors in aqueous solution should be taken into consideration when studying the host jump.
In summary, the prevalent H7N9 isolates in the outbreak in China contain the Q226L substitution in HA, which has also been shown to be important for the host jump of the H2, H3 and H5 subtypes. H7N9 viruses preferentially bind to the avian receptor but have evolved the capacity to bind to human receptors, and it is likely that they will acquire increasing human receptor-binding preference with specific amino acid substitutions in the receptor-binding site. Although airborne transmission of H7N9 in ferrets was shown to be limited76,77,78,79,90, the possibility that current H7N9 viruses might develop airborne transmissibility after repeated passage in mammals cannot be excluded.
Summary and conclusions
The determinants that contribute to the host jump of avian influenza A viruses are complex and involve several viral and host factors. Recent crystallographic studies have provided molecular insights into the shift in receptor binding caused by mutations in the viral envelope protein HA, which is a major (but not the only) determinant of the host switch. To efficiently switch, viruses must acquire a preference for the human receptor, or at least the ability to bind weakly to the human receptor in addition to the avian receptor to successfully infect and replicate in human epithelial cells in the URT, which predominantly express the α2,6-linked SA receptor. It is likely that decreased binding to avian receptors is required for human-to-human transmission. This is because the human URT is covered with secreted mucin molecules that contain the α2,3-linked SA receptor, so viruses could get trapped in the URT as these mucin molecules (containing attached virus) are tightly bound to the respiratory epithelium and are unlikely to be transmitted in droplets generated by coughing or sneezing91.
Amino acid substitutions in HA have been identified as major determinants for preferential targeting of human, rather than avian, receptors. The H1, H2 and H3 subtypes of influenza A viruses have naturally adapted to humans, causing worldwide pandemics and epidemics. In the H1 subtype, the amino acids at positions 190 and 225 in the receptor-binding sites seem to be important for the shift in receptor-binding specificity and, in addition, different combinations of mutations result in altered receptor-binding specificity: H1 HA proteins that contain E190/G225, E190/D225 or D190/G225 in their receptor-binding site have dual receptor-binding specificity, whereas those that contain the D190/D225 and D190/E225 substitutions specifically bind to the human receptor. The Q226L and G228S substitutions in the HA glycoproteins of H2 and H3 subtypes are sufficient to change the receptor-binding preference from the avian receptor to the human receptor. Moreover, experimental adaptation of the H5 subtype showed that the Q226L substitution and loss of a glycosylation site near the receptor-binding site contribute to the shift in receptor-binding preference from avian to human. Finally, in the H7 subtype, amino acid substitutions at positions 186 and 226 increase binding to the human receptor; however, H7 HA still preferentially binds to the avian receptor, and the amino acid substitutions that are responsible for the shift in receptor-binding specificity remain to be determined.
Although the molecular bases of the receptor-binding preference shifts for H1, H2, H3 and H5 HA proteins have been established, the mechanism for other HA subtypes remains unknown, particularly for H7 and H9. In the future, more efforts are needed to elucidate the molecular basis of the host jump of the other HA subtypes, which should aid the rapid identification of newly emerging epidemic and pandemic strains. Furthermore, owing to technical limitations, we can currently only solve the structure of HA in complex with simple receptor analogues with pentasaccharide or trisaccharide. However, the sialylated glycan receptors in different hosts and tissues are far more complex, with different oligosaccharide chains in the receptor, and the molecular interaction between HA and complex sialylated glycan receptors should be studied in the future.
In conclusion, as single amino acid changes seem to be sufficient to alter receptor-binding preference, and as natural selection is unpredictable, extensive surveillance of influenza viruses will be crucial for the prevention and control of future pandemics.
References
Taubenberger, J. K. & Morens, D. M. Pandemic influenza — including a risk assessment of H5N1. Rev. Sci. Tech. 28, 187–202 (2009).
Wu, Y. & Gao, G. F. Lessons learnt from the human infections of avian-origin influenza A H7N9 virus: live free markets and human health. Sci. China Life Sci. 56, 493–494 (2013).
Liu, X., Zhao, Z. & Liu, W. Insights into the roles of cyclophilin A during influenza virus infection. Viruses 5, 182–191 (2013).
Wu, Y., Wu, Y., Tefsen, B., Shi, Y. & Gao, G. F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 22, 183–191 (2014).
Muramoto, Y., Noda, T., Kawakami, E., Akkina, R. & Kawaoka, Y. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87, 2455–2462 (2013).
Gamblin, S. J. & Skehel, J. J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 285, 28403–28409 (2010).
Tong, S. et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA 109, 4269–4274 (2012).
Tong, S. et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 9, e1003657 (2013).
Air, G. M. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc. Natl Acad. Sci. USA 78, 7639–7643 (1981).
Nobusawa, E. et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182, 475–485 (1991).
Medina, R. A. & Garcia-Sastre, A. Influenza A viruses: new research developments. Nature Rev. Microbiol. 9, 590–603 (2011).
de Graaf, M. & Fouchier, R. A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 33, 823–841 (2014).
Wiley, D. C. & Skehel, J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56, 365–394 (1987).
Huang, R. T., Rott, R. & Klenk, H. D. Influenza viruses cause hemolysis and fusion of cells. Virology 110, 243–247 (1981).
Maeda, T. & Ohnishi, S. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 122, 283–287 (1980).
White, J., Matlin, K. & Helenius, A. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89, 674–679 (1981).
Shaw, M. L., Stone, K. L., Colangelo, C. M., Gulcicek, E. E. & Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 4, e1000085 (2008).
Leser, G. P. & Lamb, R. A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342, 215–227 (2005).
Sauter, N. K. et al. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry 28, 8388–8396 (1989).
Takemoto, D. K., Skehel, J. J. & Wiley, D. C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217, 452–458 (1996).
Gambaryan, A. S. et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology 232, 345–350 (1997).
Eisen, M. B., Sabesan, S., Skehel, J. J. & Wiley, D. C. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 232, 19–31 (1997).
Chu, V. C. & Whittaker, G. R. Influenza virus entry and infection require host cell N-linked glycoprotein. Proc. Natl Acad. Sci. USA 101, 18153–18158 (2004).
de Vries, E. et al. Influenza A virus entry into cells lacking sialylated N-glycans. Proc. Natl Acad. Sci. USA 109, 7457–7462 (2012).
Ablan, S., Rawat, S. S., Blumenthal, R. & Puri, A. Entry of influenza virus into a glycosphingolipid-deficient mouse skin fibroblast cell line. Arch. Virol. 146, 2227–2238 (2001).
Shinya, K. et al. Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 (2006).
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373 (1981). This paper provides the first three-dimensional structure of influenza virus HA.
Shi, Y. et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342, 243–247 (2013).
Xu, R., McBride, R., Nycholat, C. M., Paulson, J. C. & Wilson, I. A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 86, 982–990 (2012).
Zhang, W. et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J. Virol. 87, 5949–5958 (2013).
Gamblin, S. J. et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303, 1838–1842 (2004). References 29, 30 and 31 provide a structural explanation for the receptor-binding shift of H1 subtype viruses.
Liu, J. et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl Acad. Sci. USA 106, 17175–17180 (2009). This paper provides the structural basis of the receptor-binding shift of H2 subtype viruses.
Lin, T. et al. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology 392, 73–81 (2009).
Alonso, M. et al. Resistance and virulence mutations in patients with persistent infection by pandemic 2009 A/H1N1 influenza. J. Clin. Virol. 50, 114–118 (2011).
Reid, A. H., Fanning, T. G., Hultin, J. V. & Taubenberger, J. K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl Acad. Sci. USA 96, 1651–1656 (1999).
Rogers, G. N. & D'Souza, B. L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173, 317–322 (1989).
Cox, N. J. & Subbarao, K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51, 407–421 (2000).
Pappas, C. et al. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS ONE 5, e11158 (2010).
Matrosovich, M. et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74, 8502–8512 (2000).
Ha, Y., Stevens, D. J., Skehel, J. J. & Wiley, D. C. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309, 209–218 (2003).
Lin, Y. P. et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl Acad. Sci. USA 109, 21474–21479 (2012). References 40 and 41 describe the structural basis of the receptor-binding shift of H3 subtype viruses.
Nobusawa, E., Ishihara, H., Morishita, T., Sato, K. & Nakajima, K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology 278, 587–596 (2000).
Stevens, J. et al. Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J. Virol. 84, 8287–8299 (2010).
Owen, R. E. et al. Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J. Virol. 81, 11170–11178 (2007).
Asaoka, N. et al. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect. 8, 511–519 (2006).
[No authors listed.] Isolation of avian influenza A(H5N1) viruses from humans — Hong Kong, May–December 1997. MMWR Morb. Mortal. Wkly Rep. 46, 1204–1207 (1997).
Stevens, J. et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J. Mol. Biol. 381, 1382–1394 (2008).
Watanabe, Y. et al. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 7, e1002068 (2011).
Gao, Y. et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5, e1000709 (2009).
Herfst, S. et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012).
Imai, M. et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012).
Chen, L. M. et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422, 105–113 (2012).
Zhang, W. et al. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science 340, 1463–1467 (2013).
Xiong, X. et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497, 392–396 (2013). References 53 and 54 describe the structural basis of the receptor-binding shift in artificial avian influenza H5N1 viruses that have been modified to be transmissible through the air.
Lu, X. et al. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4, 502–511 (2013).
Stevens, J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006).
Ha, Y., Stevens, D. J., Skehel, J. J. & Wiley, D. C. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl Acad. Sci. USA 98, 11181–11186 (2001).
Linster, M. et al. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157, 329–339 (2014).
Zhang, Y. et al. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 340, 1459–1463 (2013).
Maher, J. A. & DeStefano, J. The ferret: an animal model to study influenza virus. Lab Anim. 33, 50–53 (2004).
Russell, C. A. et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336, 1541–1547 (2012).
Webster, R. G., Geraci, J., Petursson, G. & Skirnisson, K. Conjunctivitis in human beings caused by influenza A virus of seals. N. Engl. J. Med. 304, 911 (1981).
Banks, J., Speidel, E. & Alexander, D. J. Characterisation of an avian influenza A virus isolated from a human — is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch. Virol. 143, 781–787 (1998).
[No authors listed.] Update: influenza activity — United States and worldwide, 2003–2004 season, and composition of the 2004–2005 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 53, 547–552 (2004).
Fouchier, R. A. et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl Acad. Sci. USA 101, 1356–1361 (2004).
Hirst, M. et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 10, 2192–2195 (2004).
Koopmans, M. et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363, 587–593 (2004).
Kurtz, J., Manvell, R. J. & Banks, J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet 348, 901–902 (1996).
Nguyen-Van-Tam, J. S. et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 11, E060504.2 (2006).
Puzelli, S. et al. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J. Infect. Dis. 192, 1318–1322 (2005).
Tweed, S. A. et al. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 10, 2196–2199 (2004).
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013).
Chen, Y. et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–1925 (2013).
Li, J. et al. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Sci. China Life Sci. 56, 485–492 (2013).
Bao, C. J. et al. Live-animal markets and influenza A (H7N9) virus infection. N. Engl. J. Med. 368, 2337–2339 (2013).
Zhu, H. et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341, 183–186 (2013).
Belser, J. A. et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501, 556–559 (2013).
Watanabe, T. et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501, 551–555 (2013).
Richard, M. et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501, 560–563 (2013).
Xiong, X. et al. Receptor binding by an H7N9 influenza virus from humans. Nature 499, 496–499 (2013).
Xu, R. et al. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342, 1230–1235 (2013).
Yang, H., Carney, P. J., Chang, J. C., Villanueva, J. M. & Stevens, J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J. Virol. 87, 12433–12446 (2013). References 28, 80, 81 and 82 describe the receptor-binding properties of 2013 human-infecting H7N9 influenza viruses and the structural basis for the host jump.
Cui, L. et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nature Commun. 5, 3142 (2014).
Breg, J., Kroon-Batenburg, L. M., Strecker, G., Montreuil, J. & Vliegenthart, J. F. Conformational analysis of the sialyl α(2→3/6)N-acetyllactosamine structural element occurring in glycoproteins, by two-dimensional NOE 1H-NMR spectroscopy in combination with energy calculations by hard-sphere exo-anomeric and molecular mechanics force-field with hydrogen-bonding potential. Eur. J. Biochem. 178, 727–739 (1989).
Poppe, L., Stuike-Prill, R., Meyer, B. & van Halbeek, H. The solution conformation of sialyl-α(2→6)-lactose studied by modern NMR techniques and Monte Carlo simulations. J. Biomol. NMR 2, 109–136 (1992).
Sabesan, S., Bock, K. & Paulson, J. C. Conformational analysis of sialyloligosaccharides. Carbohydr. Res. 218, 27–54 (1991).
Stehle, T. & Harrison, S. C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 4, 183–194 (1996).
Wright, C. S. & Jaeger, J. Crystallographic refinement and structure analysis of the complex of wheat germ agglutinin with a bivalent sialoglycopeptide from glycophorin A. J. Mol. Biol. 232, 620–638 (1993).
Merritt, E. A. et al. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 3, 166–175 (1994).
Lam, T. T. et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502, 241–244 (2013).
Couceiro, J. N., Paulson, J. C. & Baum, L. G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29, 155–165 (1993).
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179 (1992).
Wise, H. M. et al. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83, 8021–8031 (2009).
Jagger, B. W. et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337, 199–204 (2012).
Acknowledgements
Work in the authors' laboratory was supported by the China Ministry of Science and Technology National 973 Project (grant no. 2011CB504703), the National Natural Science Foundation of China (NSFC; grant no. 81290342 and 31402196) and intramural special grant for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L09). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (grant no. 81321063). Y.S. is supported by the Excellent Young Scientist Program of the Chinese Academy of Sciences. The authors are grateful to B. Tefsen and J. Haywood for their help in preparing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Herd immunity
-
(Also known as herd effect or community immunity). A form of immunity that occurs when the vaccination of a substantial portion of a population (or herd) provides a measure of protection for individuals who have not developed immunity.
- Macropinocytic endocytosis
-
A process by which cells internalize molecules using a non-selective uptake pattern.
- Clathrin-mediated endocytosis
-
A process by which cells internalize molecules in a receptor-mediated uptake pattern; it is also called receptor-mediated endocytosis.
- Trans conformation
-
The conformation of a glycosidic linkage of receptor analogues, in which the two carbon atoms at either end of the glycosidic bond are oriented in opposing directions.
- Cis conformation
-
The conformation of a glycosidic linkage of a receptor analogue, in which the two carbon atoms at either end of the glycosidic bond are oriented in the same direction.
- Surface plasmon resonance
-
A method used to measure the interaction of macromolecules at a surface via changes in the refractive index.
- Microarray
-
A two-dimensional array on a solid substrate (usually a glass or plastic slide) that numerous biomaterials are attached to in a regular pattern for biochemical or genetic analysis.
- Reassortant
-
A term that usually refers to a virus that contains two or more segments of nucleic acid (segmented genome) that are derived from different viruses.
Rights and permissions
About this article
Cite this article
Shi, Y., Wu, Y., Zhang, W. et al. Enabling the 'host jump': structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol 12, 822–831 (2014). https://doi.org/10.1038/nrmicro3362
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3362
This article is cited by
-
Surveillance and characterization of avian-origin H3N2 canine influenza viruses in 2021 in China
One Health Advances (2024)
-
Genetic characterization and pathogenicity of a Eurasian avian-like H1N1 swine influenza reassortant virus
Virology Journal (2022)
-
An epitope-optimized human H3N2 influenza vaccine induces broadly protective immunity in mice and ferrets
npj Vaccines (2022)
-
Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes
Nature Communications (2022)
-
Blood group-gut microbiome—health axis gains further support from landmark multi-omics study in swines
Science China Life Sciences (2022)