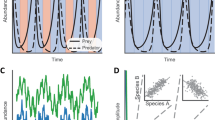
Key Points
-
Microorganisms form various ecological relationships, ranging from mutualism to competition, that in addition to other factors (such as niche preferences and random processes) shape microbial abundances. Recently, network inference techniques have frequently been applied to microbial presence–absence or abundance data to detect significant patterns of co-presence and mutual exclusion between taxa and to represent them as a network.
-
In addition to predicting links between taxa and between environmental traits and taxa, the analysis of microbial association networks reveals niches, points out keystone species and indicates alternative community configurations.
-
However, several pitfalls in the construction and interpretation of these networks exist, ranging from data normalization to multiple test correction. Thorough evaluation is needed to determine the best-performing network inference technique.
-
Recent advances in the cultivation of unknown microorganisms, combinatorial labelling and parallel cultivation may soon allow systematic co-culturing and perturbation (that is, species removal) experiments.
-
Interaction strengths that have been obtained from static networks or that have been measured experimentally can serve as inputs for dynamic models of microbial communities, which in turn can simulate the behaviour of the system in various conditions. In the long run, dynamic models could help to engineer microbial communities.
-
The theory of dynamic systems can contribute to our understanding of microbial communities. For instance, alternative community states can arise as a consequence of system dynamics without being driven by environmental differences.
Abstract
Metagenomics and 16S pyrosequencing have enabled the study of ecosystem structure and dynamics to great depth and accuracy. Co-occurrence and correlation patterns found in these data sets are increasingly used for the prediction of species interactions in environments ranging from the oceans to the human microbiome. In addition, parallelized co-culture assays and combinatorial labelling experiments allow high-throughput discovery of cooperative and competitive relationships between species. In this Review, we describe how these techniques are opening the way towards global ecosystem network prediction and the development of ecosystem-wide dynamic models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lidicker, W. Z. A. Clarification of interactions in ecological systems. Bioscience 29, 475–477 (1979).
Rodríguez-Martínez, J. M. & Pascual, A. Antimicrobial resistance in bacterial biofilms. Rev. Med. Microbiol. 17, 65–75 (2006).
Woyke, T. et al. Symbiotic insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955 (2006).
Leschine, S. B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 49, 399–426 (1995).
Gause, G. F. The Struggle for Existence (Williams & Wilkins, 1934).
Raes, J., Foerstner, K. U. & Bork, P. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr. Opin. Microbiol. 10, 1–9 (2007).
Dubelaar, G. B. J. & Jonker, R. R. Flow cytometry as a tool for the study of phytoplankton. Sci. Mar. 64, 135–156 (2000).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, 1556–1573 (2007).
Lane, D. J. et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA 82, 6955–6959 (1985).
Hamady, M. & Knight, R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19, 1141–1152 (2009).
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Cole, J. R. et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Gonzalez, A. & Knight, R. Advancing analytical algorithms and pipelines for billions of microbial sequences. Curr. Opin. Biotechnol. 23, 64–71 (2012).
De Smet, R. & Marchal, K. Advantages and limitations of current network inference methods. Nature Rev. Microbiol. 8, 717–729 (2010).
Veiga, D. F. T., Dutta, B. & Balázsi, G. Network inference and network response identification: moving genome-scale data to the next level of biological discovery. Mol. Biosyst. 6, 469–480 (2010).
Bonneau, R. et al. A predictive model for transcriptional control of physiology in a free living cell. Cell 131, 1354–1365 (2007).
Szklarczyk, D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 (2011).
Milns, I., Beale, C. M. & Smith, V. A. Revealing ecological networks using Bayesian network inference algorithms. Ecology 91, 1892–1899 (2010).
Agrawal, R., Imielinski, T. & Swami, A. Mining association rules between sets of items in large databases. ACM SIGMOD Record 22, 207–216 (1993).
Chaffron, S., Rehrauer, H., Pernthaler, J. & Von Mering, C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959 (2010).
Eiler, A., Heinrich, F. & Bertilsson, S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 6, 330–342 (2012).
Steele, J. A. et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425 (2011).
Fuhrman, J. A. Microbial community structure and its functional implications. Nature 459, 193–199 (2009).
Falkowski, P. G., Fenchel, T. & Delong, E. F. The microbial engines that drive earth's biogeochemical cycles. Science 320, 1034–1039 (2008).
Hecker, M., Lambeck, S., Toepfer, S., van Someren, E. & Guthke, R. Gene regulatory network inference: data integration in dynamic models—a review. Biosystems 96, 86–103 (2009).
May, R. M. Stability and Complexity in Model Ecosystems (Princeton Univ. Press, 1973).
Mounier, J. et al. Microbial interactions within a cheese microbial community. Appl. Environ. Microbiol. 74, 172–181 (2008).
Hoffmann, K. H. et al. Power law rank-abundance models for marine phage communities. FEMS Microbiol. Lett. 273, 224–228 (2007).
Thingstad, T. F. Elements of a theory for the mechanisms controlling abundance, diversity and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328 (2000).
Rodriguez-Brito, B. et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 4, 739–751 (2010).
Barberán, A., Bates, S. T., Casamayor, E. O. & Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351 (2012).
Zhou, J. et al. Functional molecular ecological networks. mBio 1, e00169–e00110 (2010).
Gilbert, J. A. et al. Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308 (2012).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Freilich, S. et al. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 38, 3857–3868 (2010).
Ruan, Q. et al. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22, 2532–2538 (2006).
Fuhrman, J. A. & Steele, J. A. Community structure of marine bacterioplankton: patterns, networks, and relationships to function aquat. Microb. Ecol. 53, 69–81 (2008).
The Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Faust, K. & Sathirapongsasuti, J. F. et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. (in the press).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Kolenbrander, P. E. et al. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66, 486–505 (2002).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108, 4680–4687 (2011).
Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS ONE 7, e39315 (2012).
Morgan, J. L., Darling, A. E. & Eisen, J. A. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS ONE 5, e10209 (2010).
Raes, J. & Bork, P. Molecular eco-systems biology: towards an understanding of community function. Nature Rev. Microbiol. 6, 693–699 (2008).
Aitchison, J. A concise guide to compositional data analysis. Laboratório de Estatística e Geoinformação [online], http://www.leg.ufpr.br/lib/exe/fetch.php/pessoais:abtmartins:a_concise_guide_to_compositional_data_analysis.pdf (2003).
Sogin, M. L. et al. Microbial diversity in the deep sea and the underexplored ''rare biosphere''. Proc. Natl. Acad. Sci. USA 103, 12115–12120 (2006).
Reshef, D. N. et al. Detecting novel associations in large data sets. Science 334, 1518–1524 (2011).
Kuczynski, J. et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nature Methods 7, 813–819 (2010).
Gotelli, N. J. Null model analysis of species co-occurrence patterns. Ecology 81, 2606–2621 (2000).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Margolin, A. A. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (Suppl. 1), S7 (2006).
Opgen-Rhein, R. & Strimmer, K. From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst. Biol. 1, 37 (2007).
Schaefer, J. & Strimmer, K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 21, 754–764 (2005).
Schaefer, J. & Strimmer, K. A. Shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 4, 32 (2005).
Freilich, S. et al. Competitive and cooperative metabolic interactions in bacterial communities. Nature Commun. 2, 589 (2011).
Brohée, S. et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 36, W444–W451 (2008).
Hu, Z. et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 37, W115–W121 (2009).
Smoot, M., Ono, K., Ruscheinski, J., Wang, P. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Lima-Mendez, G. & van Helden, J. The powerful law of the power law and other myths in network biology. Mol. Biosyst. 5, 1482–1493 (2009).
Paine, R. T. A note on trophic complexity and community stability. Am. Nat. 103, 91–93 (1969).
May, R. M. Biological populations with nonoverlapping generations: stable points, stable cycles, and chaos. Science 186, 645–647 (1974).
Becks, L., Hilker, F. M., Malchow, H., Jürgens, K. & Arndt, H. Experimental demonstration of chaos in a microbial food web. Nature 435, 1226–1229 (2005).
Harcombe, W. Novel cooperation experimentally evolved between species. Evolution 64, 2166–2172 (2010).
Kaeberlein, T., Lewis, K. & Epstein, S. S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129 (2002).
Nichols, D. et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 76, 2445–2450 (2010).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Park, J., Kerner, A., Burns, M. A. & Lin, X. N. Microdroplet-enabled highly parallel co-cultivation of microbial communities. PLoS ONE 6, e17019 (2011).
Kim, H. J., Du, W. & Ismagilov, R. F. Complex function by design using spatially pre-structured synthetic microbial communities: degradation of pentachlorophenol in the presence of Hg(II). Integr. Biol. 3, 126–133 (2011).
Holling, C. S. Resilience and stability of ecological systems. Annu. Rev. Ecol. Evol. Syst. 4, 1–23 (1973).
Ives, A. R. & Carpenter, S. R. Stability and diversity of ecosystems. Science 317, 58–62 (2007).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
Lewontin, R. C. The meaning of stability. Brookhaven Symp. Biol. 22, 13–23 (1969).
Petraitis, P. S. & Dudgeon, S. R. Detection of alternative stable states in marine communities. J. Exp. Mar. Biol. Ecol. 300, 343–371 (2004).
Connell, J. H. & Sousa, W. P. On the evidence needed to judge ecological stability or persistence. Am. Nat. 121, 789–824 (1983).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108, 4578–4585 (2011).
Fierer, N., Nemergut, D., Knight, R. & Craine, J. M. Changes through time: integrating microorganisms into the study of succession. Res. Microbiol. 161, 635–642 (2010).
Trosvik, P., Stenseth, N. C. & Rudi, K. Convergent temporal dynamics of the human infant gut microbiota. ISME J. 4, 151–158 (2010).
Rosindell, J., Hubbell, S. P. & Etienne, R. S. The unified neutral theory of biodiversity and biogeography at age ten. Trends Ecol. Evol. 26, 340–348 (2011).
West, S. A., Griffin, A. S., Gardner, A. & Diggle, S. P. Social evolution theory for microorganisms. Nature Rev. Microbiol. 4, 597–607 (2006).
Zomorrodi, A. R. & Maranas, C. D. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Comput. Biol. 8, e1002363 (2012).
Karlsson, F. H., Nookaew, I., Petranovic, D. & Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 29, 251–258 (2011).
Hastings, A. Transients: the key to long-term ecological understanding? Trends Ecol. Evol. 19, 39–45 (2004).
DeJongh, M. et al. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics 8, I39 (2007).
Hanly, T. J. & Henson, M. A. Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures. Biotechnol. Bioeng. 108, 376–385 (2011).
Curtis, T. P., Head, I. M. & Graham, D. W. Theoretical ecology for engineering biology. Environ. Sci. Technol. 37, 64A–70A (2003).
Dunham, M. J. Synthetic ecology: a model system for cooperation. Proc. Natl Acad. Sci. USA 104, 1741–1742 (2007).
Balagaddé, F. K. et al. A synthetic Escherichia coli predator–prey ecosystem. Mol. Syst. Biol. 4, 187 (2008).
Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Werner, J. J. et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc. Natl Acad. Sci. USA 22 Feb 2011 (doi:10.1073/pnas.1015676108).
Marsh, P. D. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).
Maloy, K. J. & Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Turnbaugh, P. J., Bäckhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Faith, J. J., McNulty, N. P., Rey, F. E. & Gordon, J. I. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science 333, 101–104 (2011).
Abbeele, P. V.d. et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for bacteroidetes and clostridium cluster IX. Appl. Environ. Microbiol. 76, 5237–5246 (2010).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Diamond, J. M. in Ecology and Evolution of Communities (eds Cody, M. & Diamond, J. M.) 342–444 (Harvard Univ. Press, 1975).
Connor, E. F. & Simberloff, D. The assembly of species communities: chance or competition? Ecology 60, 1132–1140 (1979).
Woodcock, S. et al. Neutral assembly of bacterial communities. FEMS Microbiol. Ecol. 62, 171–180 (2007).
Wootton, J. T. Field parameterization and experimental test of the neutral theory of biodiversity. Nature 433, 309–312 (2005).
Horner-Devine, M. C. et al. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88, 1345–1353 (2007).
Sloan, W. T. et al. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740 (2006).
Ricklefs, R. E. Community diversity: relative roles of local and regional processes. Science 235, 167–171 (1987).
Borgelt, C. & Kruse, R. in COMPSTAT 2002 — Proceedings in Computational Statistics: 15th Symposium held in Berlin, Germany, 2002 (eds Härdle, W. & Rönz, B.) 395–400 (Physica-Verlag, 2002).
Lallich, S., Teytaud, O. & Prudhomme, E. Association Rule Interestingness: Measure and Statistical Validation (eds Guillet, F. & Hamilton J. H.) (Springer, 2007).
Erdős, P. & Rényi, A. On the evolution of random graphs. Publ. Math. Inst. Hung. Acad. Sci. 5, 17–61 (1960).
Barabási, A.-L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Jeong, H., Mason, S. P., Barabási, A.-L. & Oltvai, Z. N. Lethality and centrality in protein networks. Nature 411, 41–42 (2001).
van Dongen, S. Graph clustering via a discrete uncoupling process. SIAM J. Matrix Analysis Appl. 30, 121–141 (2008).
Clauset, A., Newman, M. E. & Moore, C. Finding community structure in very large networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70, 066111 (2004).
Watts, D. J. & Strogatz, S. H. Collective dynamics of 'small-world' networks. Nature 393, 440–442 (1998).
Acknowledgements
K.F. and J.R. are supported by the Research Foundation Flanders (FWO), the Flemish agency for Innovation by Science and Technology (IWT) and the Brussels Institute for Research and Innovation. We would like to acknowledge G. Lima-Mendez, S. Chaffron and all other members of the Raes laboratory, as well as D. Gonze for helpful comments and discussions. We would also like to apologize to all authors whose work could not be included owing to space restraints. In adition, we thank our reviewers, whose criticisms and suggestions helped to improve this Review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Mutualism
-
An interaction between two species in which each species derives a benefit. Also referred to as cooperation or symbiosis by some authors; however, mutualism is preferred here because 'symbiosis' can be used in a broader sense to include all ecological relationships, and 'cooperation' can be used to designate mutualism between single organisms rather than populations.
- Niches
-
Defined by Hutchinson as the volume in which the growth rate of an organism is larger than or equal to 1, where the volume is an abstract space with axes that correspond to abiotic and biotic factors that affect the growth rate of the species.
- Network inference
-
The process of reconstructing the wiring diagram of a complex system from the behaviour of its components. For microbial communities, the goal of network inference is to predict ecological relationships between microorganisms from abundance data.
- Regression
-
Prediction of a relationship between a dependent variable (here, the abundance of a target species) and independent variables (the abundance (or abundances) of one or more independent source species, environmental traits and possibly a noise term), which are termed factors here.
- Lotka–Volterra equations
-
Equations that describe the dynamics of a prey–predator system. In their generalized form, the Lotka–Volterra equations can model the dynamics of more than two species and describe relationships other than prey–predator.
- Hypergeometric distribution
-
This distribution underlies Fisher's exact test, which is commonly used to infer networks from presence–absence data. Given the occurrences of two taxa across the samples, the test assesses the significance of the number of observed co-presences.
- Operational taxonomic unit
-
(OTU). Refers to bacterial and archaeal taxonomic groups that are derived by sequence clustering and that are thus specific to the samples analysed.
- Chaos
-
A type of dynamic behaviour that is characterized by irregular oscillations and a sensitivity to small differences in initial conditions, so that for two similar sets of start conditions, the system may behave entirely differently after some time has elapsed.
- Stable state
-
A region in (multi-dimensional) space in which the system remains and to which the system returns after a small perturbation. The stable state may be a point (also referred to as stable equilibrium or stable steady state), a limit cycle (at which the system oscillates) or may have other shapes (for example, the strange attractors of chaotic systems).
- Succession
-
The orderly and predictable manner by which communities change over time following the colonization of a new environment.
Rights and permissions
About this article
Cite this article
Faust, K., Raes, J. Microbial interactions: from networks to models. Nat Rev Microbiol 10, 538–550 (2012). https://doi.org/10.1038/nrmicro2832
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2832
This article is cited by
-
Bacterial wilt affects the structure and assembly of microbial communities along the soil-root continuum
Environmental Microbiome (2024)
-
Taxonomic dependency and spatial heterogeneity in assembly mechanisms of bacteria across complex coastal waters
Ecological Processes (2024)
-
Divergent maturational patterns of the infant bacterial and fungal gut microbiome in the first year of life are associated with inter-kingdom community dynamics and infant nutrition
Microbiome (2024)
-
Diversity of information pathways drives sparsity in real-world networks
Nature Physics (2024)
-
Biogeographic patterns and drivers of soil viromes
Nature Ecology & Evolution (2024)