Key Points
-
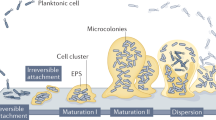
The predominant mode of growth of most bacteria in natural and engineered environments is as a surface-associated community encased in an extracellular matrix, called a biofilm. When conditions within the biofilm become unfavourable, bacteria must be able to disperse to escape and colonize new habitats.
-
The dispersal response of bacterial biofilms is regulated through the production and perception of extracellular and intracellular signalling molecules and in response to environmental cues such as changes in nutrient concentrations. Such signals and cues are translated into changes in gene expression that induce effectors, such as enzymes and surfactants, which break down the biofilm matrix and prepare bacteria for planktonic growth.
-
In addition to releasing bacteria to colonize new sites, dispersal is associated with the formation of genetic variants that may be altered in traits which are important for colonization of and competition in new habitats.
-
The sessile (biofilm) and motile (dispersal) phases of bacterial growth have close analogies to the lifestyles of colonial and holometabolous eukaryotes, including the generation of variants in the dispersal propagules. Biofilms may therefore be useful experimental tools to further explore ecological and evolutionary theories surrounding organisms with sessile and motile life phases.
Abstract
In most environments, bacteria reside primarily in biofilms, which are social consortia of cells that are embedded in an extracellular matrix and undergo developmental programmes resulting in a predictable biofilm 'life cycle'. Recent research on many different bacterial species has now shown that the final stage in this life cycle includes the production and release of differentiated dispersal cells. The formation of these cells and their eventual dispersal is initiated through diverse and remarkably sophisticated mechanisms, suggesting that there are strong evolutionary pressures for dispersal from an otherwise largely sessile biofilm. The evolutionary aspect of biofilm dispersal is now being explored through the integration of molecular microbiology with eukaryotic ecological and evolutionary theory, which provides a broad conceptual framework for the diversity of specific mechanisms underlying biofilm dispersal. Here, we review recent progress in this emerging field and suggest that the merging of detailed molecular mechanisms with ecological theory will significantly advance our understanding of biofilm biology and ecology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Labbate, M. et al. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186, 692–698 (2004).
Morgan, R., Kohn, S., Hwang, S. H., Hassett, D. J. & Sauer, K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188, 7335–7343 (2006).
Southey-Pillig, C. J., Davies, D. G. & Sauer, K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 8114–8126 (2005). This paper shows that differential protein production occurs in specific stages of biofilm development.
Tolker-Nielsen, T. et al. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182, 6482–6489 (2000).
Kjelleberg, S. & Givskov, M. in The Biofilm Mode of Life: Mechanisms and Adaptations (eds Kjelleberg, S. & Givskov, M.) 5–21 (Horizon Bioscience, 2007).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nature Rev. Microbiol. 8, 623–633 (2010).
Monds, R. D. & O'Toole, G. A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17, 73–87 (2009).
Barken, K. B. et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343 (2008).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev. Microbiol. 2, 95–108 (2004).
Periasamy, S. & Kolenbrander, P. E. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella Species in saliva. Infect. Immun. 77, 3542–3551 (2009).
Kolenbrander, P. E., Palmer, R. J., Periasamy, S. & Jakubovics, N. S. Oral multispecies biofilm development and the key role of cell–cell distance. Nature Rev. Microbiol. 8, 471–480 (2010).
Ross, P. et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281 (1987).
Merighi, M. & Lory, S. in Pseudomonas Vol. 6 (eds Ramos, J. L. & Filloux, A.) 97–138 (Springer, the Netherlands, 2010).
Webb, J. S. et al. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592 (2003).
Mai-Prochnow, A. et al. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70, 3232–3238 (2004).
Rice, S. A. et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3, 271–282 (2009). This study demonstrates that biofilm development, cell death, stability of the biofilm and the formation of dispersal variants in P. aeruginosa are dependent on the presence of a prophage in the host genome.
Purevdorj-Gage, B., Costerton, W. J. & Stoodley, P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569–1576 (2005). These authors find that the biofilms of P. aeruginosa undergo active dispersal by means of a motile subpopulation within microcolonies, and that dispersal is dependent on quorum sensing.
Barraud, N. et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353 (2006). This article shows that P. areuginosa biofilms produce low levels of NO, which acts as a signal to induce dispersal. This dispersal response is also associated with increased sensitivity to antimicrobials.
Rollet, C., Gal, L. & Guzzo, J. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 290, 135–142 (2009).
Gjermansen, M., Ragas, P., Sternberg, C., Molin, S. & Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7, 894–904 (2005). This investigation demonstrates that starvation-induced dispersal of P. putida is mediated via cleavage and release of the adhesin LapA, and that the starvation cue operates via the intracellular second messenger c-di-GMP.
Hunt, S. M., Werner, E. M., Huang, B., Hamilton, M. A. & Stewart, P. S. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70, 7418–7425 (2004).
Sauer, K. et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186, 7312–7326 (2004).
Schleheck, D. et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS Biol. 4, e5513 (2009).
An, S., Wu, J. e. & Zhang, L.-H. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76, 8160–8173 (2010).
Thormann, K. M., Saville, R. M., Shukla, S. & Spormann, A. M. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187, 1014–1021 (2005).
Barraud, N. et al. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2, 370–378 (2009).
Schlag, S., Nerz, C., Birkenstock, T. A., Altenberend, F. & Gotz, F. Inhibition of staphylococcal biofilm formation by nitrite. J. Bacteriol. 189, 7911–7919 (2007).
Kaplan, J. B. & Fine, D. H. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68, 4943–4950 (2002).
Musk, D. J., Banko, D. A. & Hergenrother, P. J. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12, 789–796 (2005).
Glick, R. et al. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 192, 2973–2980 (2010).
Moreau-Marquis, S. et al. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am. J. Physiol. Lung Cell. Mol. Physiol. 295, L25–L37 (2008).
Kaplan, J. B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. Crit. Rev. Oral Biol. Med. 89, 205–218 (2010).
Lauderdale, K. J., Malone, C. L., Boles, B. R., Morcuende, J. & Horswill, A. R. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 28, 55–61 (2010).
Rice, S. A. et al. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187, 3477–3485 (2005).
Boles, B. R. & Horswill, A. R. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 (2008).
Crossman, L. & Dow, J. M. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 6, 623–629 (2004).
Davies, D. G. & Marques, C. N. H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 (2009).
Kolodkin-Gal, I. et al. D-amino acids trigger biofilm disassembly. Science 328, 627–629, (2010). This report describes the role of D -amino acids in triggering biofilm dispersal in B. subtilis and shows that the mode of action is via triggering detachment of amyloid fibres from the membrane. In addition, it shows that D -amino acids inhibit biofilm formation by P. aeruginosa and S. aureus.
Sawyer, L. K. & Hermanowicz, S. W. Detachment of Aeromonas hydrophila and Pseudomonas aeruginosa due to variations in nutrient supply. Water Sci. Technol. 41, 139–145 (2000).
Delaquis, P. J., Caldwell, D. E., Lawrence, J. R. & Mccurdy, A. R. Detachment of Pseudomonas fluorescens from biofilms on glass surfaces in response to nutrient stress. Microb. Ecol. 18, 199–210 (1989).
Delille, A., Quiles, F. & Humbert, F. In situ monitoring of the nascent Pseudomonas fluorescens biofilm response to variations in the dissolved organic carbon level in low-nutrient water by attenuated total reflectance-fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 73, 5782–5788 (2007).
Gjermansen, M., Nilsson, M., Yang, L. & Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75, 815–826 (2010).
James, G. A., Korber, D. R., Caldwell, D. E. & Costerton, J. W. Digital image-analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177, 907–915 (1995).
Kader, A., Simm, R., Gerstel, U., Morr, M. & Römling, U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60, 602–616 (2006).
Merritt, J. H. et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1, e00183-10 (2010).
Paul, R. et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727 (2004).
Barraud, N. et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191, 7333–7342 (2009).
Carlson, H. K., Vance, R. E. & Marletta, M. A. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 77, 930–942 (2010).
Schmidt, I., Steenbakkers, P. J. M., op den Camp, H. J. M., Schmidt, K. & Jetten, M. S. M. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J. Bacteriol. 186, 2781–2788 (2004).
Potter, A. J. et al. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J. Infect. Dis. 199, 227–235 (2009).
Delgado-Nixon, V. M., Gonzalez, G. & Gilles-Gonzalez, M. A. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39, 2685–2691 (2000).
Alexandre, G. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 156, 2283–2293 (2010).
Giba, Z., Grubišić, D. & Konjević, R. in Nitric Oxide in Plant Growth, Development and Stress Physiology (Plant Cell Monographs) Vol. 5 (eds Lamattina, L. & Polacco, J.) 91–111 (Springer, Berlin, 2007).
Kumar, A., Toledo, J. C., Patel, R. P., Lancaster, J. R. Jr & Steyn, A. J. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl Acad. Sci. USA 104, 11568–11573 (2007).
Bishop, C. D. & Brandhorst, B. P. On nitric oxide signaling, metamorphosis, and the evolution of biphasic life cycles. Evol. Dev. 5, 542–550 (2003).
Davies, D. G. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998). The authors report that quorum sensing signalling is involved in the development of P. aeruginosa biofilms and that biofilms formed by quorum sensing mutants are less stable than those formed by wild-type cells.
Abee, T., Kovasc, A. T., Kuipers, O. P. & van der Veen, S. Biofilm formation and dispersal in Gram-positivie bacteria. Curr. Opin. Biotechnol. 22, 172–179 (2011).
Yarwood, J. M., Bartels, D. J., Volper, E. M. & Greenberg, E. P. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850 (2004).
Giacometti, A. et al. RNA III inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47, 1979–1983 (2003).
Orent, W. Slime city: where germs talk to each other and execute precise attacks. Discover (17 Jul 2009).
Hammer, B. K. & Bassler, B. L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–104 (2003).
Puskas, A., Greenberg, E. P., Kaplan, S. & Schaefer, A. L. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179, 7530–7537 (1997).
Boles, B. R., Thoendel, M. & Singh, P. K. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57, 1210–1223 (2005).
Jobling, M. G. & Holmes, R. K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26, 1023–1034 (1997).
Dow, J. M. et al. Biofilm dispersal in Xanthomonas campestris is controlled by cell–cell signaling and is required for full virulence to plants. Proc. Natl Acad. Sci. USA 100, 10995–11000 (2003). DSF (later chemically determined to be cis -11-methyl-2-dodecenoic acid) is shown to be important for dispersing aggregates of the plant pathogen X. campestris . Additional DSF-like compounds that regulate dispersal in other bacteria have subsequently been identified.
Ryan, R. P. et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl Acad. Sci. USA 103, 6712–6717 (2006).
Tao, F., He, Y.-W., Wu, D.-H., Swarup, S. & Zhang, L.-H. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 192, 1020–1029 (2010).
Deng, Y., Wu, J. Ä., Tao, F. & Zhang, L.-H. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem. Rev. 111, 160–173 (2011).
Huynh, T. T. Carbon starvation induced dispersal of Pseudomonas aeruginosa biofilms. Thesis, Univ. New South Wales (2011).
Lam, H. et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555 (2009).
Ortzen, D. & Nielsen, P. H. We find them here, we find them there: functional bacterial amyloid. Cell. Mol. Life Sci. 65, 910–927 (2008).
Karatan, E. & Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 (2009).
Allison, D. G., Ruiz, B., SanJose, C., Jaspe, A. & Gilbert, P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilm. FEMS Microbiol. Lett. 167, 179–184 (1998).
Baty, A. M. I., Eastburn, C. C., Techkarnjanaruk, S., Goodman, A. E. & Geesey, G. G. Spatial and temporal variations in chitinolytic gene expression and bacterial biomass production during chitin degradation. Appl. Environ. Microbiol. 66, 3574–3585 (2000).
Mann, E. E. et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4, e5822 (2009).
Manuel, S. G. A. Role of active-site residues of dispersin B, a biofilm-releasing β-hexosaminidase from a periodontal pathogen, in substrate hydrolysis. FEBS J. 274, 5987–5999 (2007).
Kaplan, J. B., Meyenhofer, M. F. & Fine, D. H. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185, 1399–1404 (2003).
Kaplan, J. B., Ragunath, C., Velliyagounder, K., Fine, D. H. & Ramasubbu, N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48, 2633–2636 (2004).
Chaignon, P. et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75, 125–132 (2007).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002). This paper describes the role of eDNA as a key component of the extracellular biofilm matrix and shows that enzymatic digestion of eDNA destabilizes P. aeruginosa biofilms.
Boyd, A. & Chakrabarty, A. M. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60, 2355–2359 (1994).
Schooling, S. R., Charaf, U. K., Allison, D. G. & Gilbert, P. A role for rhamnolipid in biofilm dispersion. Biofilms 1, 91–99 (2004).
Kuiper, I. et al. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51, 97–113 (2004).
Paul, J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2, 579–589 (2008).
Garcia-Contreras, R., Zhang, X.-S., Kim, Y. & Wood, T. K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS ONE 3, e2394 (2008).
Sillankorva, S., Neubauer, P. & Azeredo, J. Phage control of dual species biofilms of Pseudomonas fluorescens and Staphylococcus lentus. Biofouling 26, 567–575 (2010).
Whiteley, M. et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001).
Kirov, S. M. et al. Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 153, 3264–3274 (2007).
Zegans, M. E. et al. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191, 210–219 (2009).
Hughes, K. A., Sutherland, I. W. & Jones, M. V. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144, 3039–3047 (1998).
Hughes, K. A., Sutherland, I. W., Clark, J. & Jones, M. V. Bacteriophage and associated polysaccharide depolymerases: novel tools for study of bacterial biofilms. J. Appl. Microbiol. 85, 583–590 (1998).
Lu, T. K. & Collins, J. J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA 104, 11197–11202 (2007).
Mitchell, H. L. et al. Treponema denticola biofilm-induced expression of a bacteriophage, toxin–antitoxin systems and transposases. Microbiology 156, 774–788 (2010).
Bowler, D. E. & Benton, T. G. Causes and consequences of animal dispersal strategies: relating individual behaviour to spatial dynamics. Biol. Rev. 80, 205–225 (2005).
Ronce, O. How does it feel to be a rolling stone? Ten questions about dispersal evolution. Annu. Rev. Ecol. Evol. Sys. 38, 231–253 (2007). This and reference 94 review some of the key ecological and evolutionary issues in the field of dispersal, with reference 95 in particular being accessible for workers outside the field.
Clobert, J., Le Galliard, J.-F., Cote, J., Meylan, S. & Massot, M. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 12, 197–209 (2009).
Rosenberg, E., Kaplan, N., Pines, O., Rosenberg, M. & Gutnick, D. Capsular polysaccharides interfere with adherence of Acinetobacter calcoaceticus to hydrocarbon. FEMS Microbiol. Lett. 17, 157–160 (1983).
Hentzer, M., Eberl, L. & Givskov, M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2, 37–61 (2005).
Ito, A., May, T., Kawata, K. & Okabe, S. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol. Bioeng. 99, 1462–1471 (2008).
Banin, E., Brady, K. M. & Greenberg, E. P. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72, 2064–2069 (2006).
Rowe, M. C., Withers, H. L. & Swift, S. Uropathogenic Escherichia coli forms biofilm aggregates under iron restriction that disperse upon the supply of iron. FEMS Microbiol. Lett. 307, 102–109 (2010).
Matz, C. & Kjelleberg, S. Off the hook – how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307 (2005).
Hausner, M. & Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65, 3710–3713 (1999).
Marshall, D. J. & Keough, M. J. The evolutionary ecology of offspring size in marine invertebrates. Adv. Mar. Biol. 53, 1–60 (2007).
Strathmann, R. R. Feeding and nonfeeding larval development and life-history evolution in marine invertebrates. Annu. Rev. Ecol. Sys. 16, 339–361 (1985).
Sanchez-Contreras, M. et al. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184, 1587–1596 (2002).
Webb, J. S., Lau, M. & Kjelleberg, S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186, 8066–8073 (2004).
Koh, K. S. et al. Phenotypic diversification and adaptation of Serratia marcescens MG1 biofilm derived morphotypes. J. Bacteriol. 189, 119–130 (2007). This work uncovers the temporally regulated production of morphotypic variants during biofilm development in S. marcescens , and shows that such variants are phentoypically distinct from wild-type cells and also differ in biofilm development.
Koh, K. S. et al. Minimal genetic diversity enhances predation resistance. Mol. Ecol. (in the press).
Benach, J. et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26, 5153–5166 (2007).
Christen, M. et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl Acad. Sci, USA 104, 4112–4117 (2007).
Hickman, J. W. & Harwood, C. S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 (2008).
Lee, V. T. et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484 (2007).
Ma, Q., Yang, Z., Pu, M., Peti, W. & Wood, T. K. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ. Microbiol. 13, 631–642 (2011).
Landini, P., Antoniani, D., Burgess, J. & Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 86, 813–823 (2010).
Izano, E. A., Amarante, M. A., Kher, W. B. & Kaplan, J. B. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476 (2008).
Elton, C. S. The Ecology of Invasions by Animals and Plants (The University of Chicago Press, Chicago, 1950).
Thorson, G. Reproductive and larval ecology of marine bottom invertebrates. Biol. Rev. 25, 1–45 (1950).
Hughes, R. N. A Functional Biology of Clonal Animals (Chapman and Hall, New York, 1989).
Jackson, J. B. C., Buss, L. W. & Cook, R. E. Population Biology and Evolution of Clonal Organisms (Yale Univ. Press, New Haven, 1985). This and reference 119 provide a comprehensive overview of the biology of modular organisms.
Harrison, P. L. et al. Mass spawning in tropical reef corals. Science 223, 1186–1189 (1984).
Brusca, R. C. & Brusca, G. J. Invertebrates (Sinauer Assoc., Inc., Sunderland, Massachusetts, 2003).
McHugh, D. & Rouse, G. W. Life history evolution of marine invertebrates: new views from phylogenetic systematics. Trends Ecol. Evol., 13, 182–186 (1998).
Acknowledgements
The authors acknowledge S. Longford for help with the figures and the Australian Research Council, Environmental Biotechnology Cooperative Research Centre and National Health and Medical Research Council for ongoing and long term support of this research. This is publication number 0058 of the Sydney Institute of Marine Science, Australia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
FURTHER INFORMATION
Centre for Marine bio-innovation, University of New South Wales
Singapore Centre for Environmental Life Sciences Engineering
Glossary
- Dispersal
-
The movement of an individual organism away from the parent organism or population to a new niche.
- Cell communication signals
-
Molecules that are produced and perceived by an organism. Signals are produced at a particular stage of growth under specific conditions or in response to changes in the environment. They accumulate extracellularly and are recognized by a dedicated receptor to induce a concerted response when a critical threshold has been reached. To be classed as cell communication, this response must extend beyond that which is required for metabolism or detoxification of the substance.
- Second messenger
-
An intracellular molecule (usually small and rapidly diffusible) that transmits information from a receptor to a target molecule; for example, cyclic AMP and cyclic di-GMP.
- Chemotaxis
-
The movement of cells or organisms according to chemical concentration gradients in the environment, either towards or away from the stimulus.
- Nitric oxide
-
(NO). A small, reactive gas and a universal signalling molecule in biological systems (as initially discovered in the 1970s, for its role in regulating vasodilation in mammals). In bacteria, NO is generated as a by-product of anaerobic metabolism or by NO synthases (NOSs).
- Autoinducing peptides
-
Extracellular peptides, ranging from 5 to 34 amino acids in length, that are generated by cleavage from precursor peptides and then further post-transcriptionally modified. These peptides are used by Gram-positive bacteria as cell communication signals.
- Sensor regulator
-
A protein that receives and responds to information about changes in the environment, either by binding second messengers or through phosphorylation, to induce transcriptional changes.
- Response regulator
-
The phosphorylation-dependent modulator of a two-component phosphorelay system. The partner sensor protein responds to environmental stimuli to modulate the phosphorylation status of the regulator, and the resultant phosphorylation cascade drives the response through differential expression of target genes.
- Lysogenic
-
Pertaining to a bacteriophage genome: being incorporated into the chromosome of the host bacterium, resulting in transmission to daughter bacterial cells on cell division. Lysogenic phages are referred to as prophages.
- Bet hedging
-
An evolutionary response to variable environments. In the context of dispersal, it is predicted to manifest in a number of ways, including the production of different types of dispersal cells to maximize colonization of different habitats, and spreading dispersal in time to accommodate temporally varying habitats.
- Colonial
-
Of an organism: able to form replicate, more or less identical units ('modules') via asexual means; these units then often connect physically and physiologically to form a colony. Monospecies biofilms are colonial (or modular, or clonal) in this sense.
- Holometabolous
-
Pertaining to an insect: with a life cycle in which there is a larval phase that is morphologically and ecologically distinct from the adult phase and which must undergo 'complete metamorphosis' via a pupal phase before becoming an adult. Examples include butterflies and true flies.
Rights and permissions
About this article
Cite this article
McDougald, D., Rice, S., Barraud, N. et al. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10, 39–50 (2012). https://doi.org/10.1038/nrmicro2695
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2695
This article is cited by
-
Bacterial c-di-GMP signaling gene affects mussel larval metamorphosis through outer membrane vesicles and lipopolysaccharides
npj Biofilms and Microbiomes (2024)
-
Distinct bacterial population dynamics and disease dissemination after biofilm dispersal and disassembly
The ISME Journal (2023)
-
Digestive exophagy of biofilms by intestinal amoeba and its impact on stress tolerance and cytotoxicity
npj Biofilms and Microbiomes (2023)
-
Acylated and non-acylated anthocyanins as antibacterial and antibiofilm agents
Discover Food (2023)
-
Genomic insights into the c-di-GMP signaling and biofilm development in the saprophytic spirochete Leptospira biflexa
Archives of Microbiology (2023)