Key Points
-
The anti-stress molecule dimethylsulphoniopropionate (DMSP) is made in vast quantities — about 1 billion tonnes per year — by many single-celled plankton and some algal seaweeds; when it is released by these organisms into the oceans, it is a food source for many marine bacteria. The importance of the associated catabolic biotransformations in the global sulphur cycle is all the more great because one of the products, the volatile dimethyl sulphide (DMS), has several environmental effects, ranging from the initiation of cloud cover by some of its oxidation products to its ability to act as a chemoattractant for many marine animals.
-
Recent genetic and genomic analyses of several marine bacteria have provided many insights into the mechanisms of DMSP catabolism. These insights include an unexpected amount of diversity in the enzymatic mechanisms and the regulation involved, and in the identities of the microorganisms that can degrade DMSP.
-
The dmd genes that encode enzymes of the demethylation pathway for DMSP catabolism occur in many strains of abundant marine alphaproteobacteria known as the roseobacters and also in the world's most populous group of marine bacteria, the SAR11 clade. These genes are therefore widespread in metagenomic data sets from marine environments. The demethylation pathway that was revealed by molecular genetics differed from that which had been previously predicted.
-
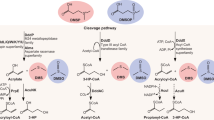
Another series of catabolic pathways involves the cleavage of DMSP by enzymes known generically as DMSP lyases, which generate dimethyl sulphide (DMS) as a primary product. A total of six enzymes, encoded by their corresponding ddd genes, were identified in different bacteria, and these differ with regard to their polypeptide families, their subcellular locations (one of them, DddY, is in the periplasm) and the identities of their catabolites. DddD generates 3-hydroxypropionate (3HP), whereas the other five — DddL, DddP, DddQ, DddW and DddY — give rise to acrylate.
-
Several ddd genes that encode different DMSP lyases are subject to horizontal gene transfer, in some cases between taxonomically diverse organisms. For example, DddP occurs not only in roseobacters, but also sporadically in other distantly related marine bacteria and, more remarkably, in some fungal pathogens.
-
Some individual bacteria have multiple ways of catabolizing DMSP. This feature is most prevalent in the roseobacters, several strains of which contain the DMSP demethylase and one or more different DMSP lyases. These different mechanisms may be adapted to particular environmental conditions.
-
In several bacteria, the regulation of DMSP cleavage is unusual, as the catabolic products, acrylate or 3HP, can act as co-inducers of the ddd genes and, hence, of the DMS-producing phenotype. Although the substrate DMSP may seem to be an effective co-inducer in some species, it must first be converted to one of the bona fide inducer molecules.
Abstract
The compatible solute dimethylsulphoniopropionate (DMSP) has important roles in marine environments. It is an anti-stress compound made by many single-celled plankton, some seaweeds and a few land plants that live by the shore. Furthermore, in the oceans it is a major source of carbon and sulphur for marine bacteria that break it down to products such as dimethyl sulphide, which are important in their own right and have wide-ranging effects, from altering animal behaviour to seeding cloud formation. In this Review, we describe how recent genetic and genomic work on the ways in which several different bacteria, and some fungi, catabolize DMSP has provided new and surprising insights into the mechanisms, regulation and possible evolution of DMSP catabolism in microorganisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Stefels, J., Steinke, M., Turner, S., Malin, G. & Belviso, S. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 83, 245–275 (2007).
Sunda, W., Kieber, D. J., Kiene, R. P. & Huntsman, S. An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320 (2002).
Wolfe, G. V., Steinke, M. & Kirst, G. O. Grazing-activated chemical defence in a unicellular marine alga. Nature 387, 894–897 (1997).
Otte, M. L., Wilson, G., Morris, J. T. & Moran, B. M. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J. Exp. Bot. 55, 1919–1925 (2004).
Malmstrom, R. R., Kiene, R. P., Cottrell, M. T. & Kirchman, D. L. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70, 4129–4135 (2004).
Vila, M. et al. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70, 4648–4657 (2004).
Tripp, H. J. et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744 (2008).
Yoch, D. C. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethyl sulfide. Appl. Environ. Microbiol. 68, 5804–5815 (2002).
Cantoni, G. L. & Anderson, D. G. Enzymatic cleavage of dimethylpropiothetin by Polysiphonia lanosa. J. Biol. Chem. 222, 171–177 (1956).
Steinke, M., Wolfe, G. V. & Kirst, G. O. Partial characterisation of dimethylsulfoniopropionate (DMSP) lyase isozymes in 6 strains of Emiliania huxleyi. Mar. Ecol. Prog. Ser. 175, 215–225 (1998).
Yost, D. M. & Mitchelmore, C. L. Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga Symbiodinium microadriaticum. Mar. Ecol. Prog. Ser. 386, 61–70 (2009).
Kiene, R. P. & Taylor, B. F. Biotransformations of organosulfur compounds in sediments via 3-mercaptopropionate. Nature 332, 148–150 (1988).
Wagner, C. & Stadtman, E. R. Bacterial fermentation of dimethyl-β-propiothetin. Arch. Biochem. Biophys. 98, 331–336 (1962).
Newton, R. J. et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798 (2010).
Ansede, J. H., Pellechia, P. J. & Yoch, D. C. Metabolism of acrylate to β-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl. Environ. Microbiol. 65, 5075–5081 (1999).
Todd, J. D. et al. Molecular dissection of bacterial acrylate catabolism – unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343 (2010). The first molecular genetics-based description of the genes and enzymes involved in an entire pathway for DMSP catabolism.
Todd, J. D. et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007). The first description of an enzyme, DddD, that cleaves DMSP to yield DMS.
Curson, A. R. J., Rogers, R., Todd, J. D., Brearley, C. A. & Johnston, A. W. B. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 757–767 (2008).
Curson, A. R. J., Sullivan, M. J., Todd, J. D. & Johnston, A. W. B. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5, 1191–1200 (2011).
Todd, J. D., Curson, A. R., Dupont, C. L., Nicholson, P. & Johnston, A. W. B. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ. Microbiol. 11, 1376–1385 (2009).
Todd, J. D. et al. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438 (2010).
Todd, J. D., Kirkwood, M., Newton-Payne, S. & Johnston, A. W. B. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 16 Jun 2011 (doi:10.1038/ismej.2011.79).
Kirkwood, M., Le Brun, N. E., Todd, J. D. & Johnston, A. W. B. The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide. Microbiology 156, 1900–1906 (2010).
Wang, Y. et al. Study on the creatinase from Paracoccus sp. strain WB1. Process Biochem. 41, 2072–2077 (2006).
Yoch, D. C., Ansede, J. H. & Rabinowitz, K. S. Evidence for intracellular and extracellular dimethylsulfoniopropionate (DMSP) lyases and DMSP uptake sites in two species of marine bacteria. Appl. Environ. Microbiol. 63, 4625–4625 (1997).
deSouza, M. P. & Yoch, D. C. in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds (eds Kiene, R. P., Visscher, P. T., Keller, M. D. & Kirst, G. O.) 293–304 (Plenum, New York, 1996).
Howard, E. C. et al. Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652 (2006). The characterization of dmdA , the first gene to be identified that is involved in DMSP catabolism, and which encodes DMSP demethylase in roseobacters and the SAR11 clade.
Reisch, C. R., Moran, M. A. & Whitman, W. B. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190, 8018–8024 (2008).
Cosquer, A. et al. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl. Environ. Microbiol. 65, 3304–3311 (1999).
Sun, L., Curson, A. R. J., Todd, J. D. & Johnston, A. W. B. Diversity of DMSP transport in marine bacteria, revealed by genetic analyses. Biogeochemistry (in the press).
Kiene, R. P., Linn, J. P. & Bruton, J. A. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–224 (2000).
Elssner, T., Engemann, C., Baumgart, K. & Kleber, H. P. Involvement of coenzyme A esters and two new enzymes, an enoyl-CoA hydratase and a CoA-transferase, in the hydration of crotonobetaine to L-carnitine by Escherichia coli. Biochemistry 40, 11140–11148 (2001).
van der Maarel, M. J. E. C., Aukema, W. & Hansen, T. A. Purification and characterization of a dimethylsulfoniopropionate cleaving enzyme from Desulfovibrio acrylicus. FEMS Microbiol. Lett. 143, 241–245 (1996).
Gross, R., Simon, J. & Kröger, A. Periplasmic methacrylate reductase activity in Wolinella succinogenes. Arch. Microbiol. 176, 310–313 (2001).
Mikoulinskaia, O., Akimenko, V., Galouchko, A., Thauer, R. K. & Hedderich, R. Cytochrome c-dependent methacrylate reductase from Geobacter sulfurreducens AM-1. Eur. J. Biochem. 263, 346–352 (1999).
Sullivan, M. J. et al. Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides. PLoS ONE 6, e15972 (2011).
Reisch, C. R. et al. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473, 208–211 (2011). The demonstration that the downstream part of the DMSP demethylation pathway is widespread in roseobacters and other bacteria.
Maddocks, S. E. & Oyston, P. C. F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623 (2008).
Bürgmann, H. et al. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ. Microbiol. 9, 2742–2755 (2007).
Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002).
Oh, H. M. et al. Complete genome sequence of “Candidatus Puniceispirillum marinum” IMCC1322, a representative of the SAR116 clade in the Alphaproteobacteria. J. Bacteriol. 192, 3240–3241 (2010).
Thrash, J. C. et al. Genome sequences of strains HTCC2148 and HTCC2080, belonging to the OM60/NOR5 clade of the gammaproteobacteria. J. Bacteriol. 192, 3842–3843 (2010).
Tang, K., Huang, H., Jiao, N. & Wu, C. H. Phylogenomic analysis of marine Roseobacters. PLoS ONE 5, e11604 (2010).
Raina, J. B., Tapiolas, D., Willis, B. L. & Bourne, D. G. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol 75, 3492–3501 (2009).
Curson, A. R. J., Fowler, E. K., S. Dickens, Johnston, A. W. B. & Todd, J. D. Multiple DMSP lyases in the γ-proteobacterium Oceanimonas doudoroffii. Biogeochemistry (in the press).
Kirkwood, M., Todd, J. D., Rypien, K. L. & Johnston, A. W. The opportunistic coral pathogen Aspergillus sydowii contains dddP and makes dimethyl sulfide from dimethylsulfoniopropionate. ISME J. 4, 147–150 (2010).
Bacic, M. K. & Yoch, D. C. In vivo characterization of dimethylsulfoniopropionate lyase in the fungus Fusarium lateritium. Appl. Environ. Microbiol. 64, 106–111 (1998).
Ansede, J. H., Friedman, R. & Yoch, D. C. Phylogenetic analysis of culturable dimethyl sulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl. Environ. Microbiol. 67, 1210–1217 (2001).
Curson, A. R., Sullivan, M. J., Todd, J. D. & Johnston, A. W. B. Identification of genes for dimethyl sulfide production in bacteria in the gut of Atlantic Herring (Clupea harengus). ISME J. 4, 144–146 (2010).
Trinick, M. J. Symbiosis between Rhizobium and the non-legume, Trema aspera. Nature 244, 459–460 (1973).
Ramette, A., LiPuma, J. J. & Tiedje, J. M. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71, 1193–1201 (2005).
Dickschat, J. S., Zell, C. & Brock, N. L. Pathways and substrate specificity of DMSP catabolism in marine bacteria of the Roseobacter clade. Chembiochem 15, 417–425 (2010).
Howard, E. C., Sun, S., Biers, E. J. & Moran, M. A. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10, 2397–2410 (2008).
Raina, J. B., Dinsdale, E. A., Willis, B. L. & Bourne, D. G. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18, 101–108 (2010).
Rusch, D. B. et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77 (2007). One of a series of papers from the J. Craig Venter Institute that describes an impressive marine-metagenomics database that can be interrogated for any gene of interest.
Gifford, S. M., Sharma, S., Rinta-Kanto, J. M. & Moran, M. A. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5, 461–472 (2011).
Vila-Costa, M. et al. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4, 1410–1420 (2010).
Rinta-Kanto, J. M. et al. Analysis of sulfur-related transcription by Roseobacter communities using a taxon-specific functional gene microarray. Environ. Microbiol. 13, 453–467 (2011).
Chen, Y. & Murrell, J. C. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 18, 157–163 (2010).
Mou, X., Sun, S., Edwards, R. A., Hodson, R. E. & Moran, M. A. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451, 708–711 (2008).
Gonzalez, J. M., Kiene, R. P. & Moran, M. A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65, 3810–3819 (1999).
Dai, Y., Pochapsky, T. C. & Abeles, R. H. Mechanistic studies of two dioxygenases in the methionine salvage pathway of Klebsiella pneumoniae. Biochemistry 40, 6379–6387 (2001).
Boden, R., Kelly, D. P., Murrell, J. C. & Schäfer, H. Oxidation of dimethylsulfide to tetrathionate by Methylophaga thiooxidans sp. nov.: a new link in the sulfur cycle. Environ. Microbiol. 12, 2688–2699 (2010).
Schäfer, H., Myronova, N. & Boden, R. Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J. Exp. Bot. 61, 315–334 (2010).
Lovelock, J., Maggs, R. & Rasmussen, R. Atmospheric dimethyl sulfide and the natural sulfur cycle. Nature 237, 452–453 (1972).
Charlson, R. J., Lovelock, J. E., Andreae, M. O. & Warren, S. G. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326, 655–661 (1987).
Vallina, S. M. & Simo, R. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315, 506–508 (2007).
DeBose, J. L. & Nevitt, G. A. The use of odors at different spatial scales: comparing birds with fish. J. Chem. Ecol. 34, 867–881 (2008).
Nevitt, G. A. Sensory ecology on the high seas: the odor world of the procellariiform seabirds. J. Exp. Biol. 211, 1706–1713 (2008).
Steinke, M., Stefels, J. & Stamhuis, E. Dimethyl sulfide triggers search behavior in copepods. Limnol. Oceanogr. 51, 1925–1930 (2006).
Ziegler, C., Bremer, E. & Kramer, R. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78, 13–34 (2010).
Baliarda, A., Robert, H., Jebbar, M., Blanco, C. & Le Marrec, C. Isolation and characterization of ButA, a secondary glycine betaine transport system operating in Tetragenococcus halophila. Curr. Microbiol. 47, 347–351 (2003).
Rees, D. C., Johnson, E. & Lewinson, O. ABC transporters: the power to change. Nature Rev. Mol. Cell. Biol. 10, 218–227 (2009).
Kempf, B. & Bremer, E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330 (1998).
Reisch, C. R., Moran, M. A. & Whitman, W. B. Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2, 172 (2011).
Acknowledgements
The authors are grateful to M. Kirkwood, E. Fowler, C. Brearley, Y. Chan, L. Sun, S. Newton-Payne, N. Nikolaidou-Katsaridou and R. Green for helpful discussions. This work was funded by the UK Biotechnology and Biological Sciences Research Council and the UK National Environment Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Zwitterion
-
A molecule that contains both a positive and a negative charge at different regions in the molecule.
- Biotransformations
-
Conversions or modifications of starting compounds to form other compounds.
- Osmotolerance
-
The ability of an organism or cell to tolerate high concentrations of solutes.
- Albedo
-
A measure of the proportion of radiation that is reflected from an object in comparison with the amount that initially reaches that object. Thus, clouds reflect sunlight such that the Earth's overall albedo is ∼35%.
- Chemoattractant
-
A compound, often a nutrient, towards which motile organisms (including bacteria, animals and protists) move.
- Transcriptional regulator
-
A polypeptide that affects the level of expression of a gene, usually by binding to cis-acting DNA sequences near the promoter of that gene.
Rights and permissions
About this article
Cite this article
Curson, A., Todd, J., Sullivan, M. et al. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol 9, 849–859 (2011). https://doi.org/10.1038/nrmicro2653
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2653
This article is cited by
-
The biogeochemistry of marine dimethylsulfide
Nature Reviews Earth & Environment (2023)
-
Strong chemotaxis by marine bacteria towards polysaccharides is enhanced by the abundant organosulfur compound DMSP
Nature Communications (2023)
-
Structure and proposed DNA delivery mechanism of a marine roseophage
Nature Communications (2023)
-
Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria
The ISME Journal (2023)
-
Genome analysis of a coral-associated bacterial consortium highlights complementary hydrocarbon degradation ability and other beneficial mechanisms for the host
Scientific Reports (2023)