Key Points
-
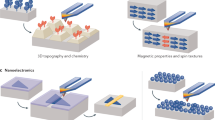
Microtechnology — the science of structures with micron or submicron-scale features — is beginning to have an impact on microbiology. Microstructured materials provide a range of capabilities for studying microorganisms because their dimensions match the intrinsic size of cells or collections of cells. Broadly stated, this class of structures offers biologists the ability to control the interface between cells and their chemical and physical environment.
-
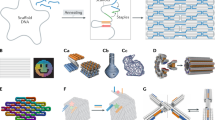
Soft lithography is a set of techniques that makes it possible to create and replicate microstructures in various materials that are biocompatible and applicable to the study of microorganisms. These techniques are easy to learn, inexpensive, and are available to microbiologists. Recent advances in soft lithography make it possible to create and replicate structures using standard equipment found in most laboratories.
-
Public foundries at Harvard, Stanford, Caltech and the University of Washington (USA), provide access to equipment for microfabrication and function as mechanisms for disseminating new techniques. Foundries teach scientists the techniques of soft lithography and provide custom-designed microfabricated materials to users at a reasonable price.
-
Soft microstructures have only recently been applied to the study of microorganisms. Examples of applications of these materials to microbiology include: the detection of pathogens, the study of interactions between microbes, microbial culture and isolation, single-cell biochemistry and genetics, and studies on motility, chemotaxis, phototaxis, quorum sensing and population dynamics.
-
Microstructures will have a crucial role in the development of new techniques for studying microorganisms. We believe that these structures will provide new capabilities for improvements in cell culture, the study of bacterial physiology and behavior, and quantitative microbiology. Soft lithographic methods are the most widely used techniques for producing classes of structures in materials that might be of interest to microbiologists.
-
Microfabrication has much to offer microbiology and soft lithography provides a bridge between these two fields. The application of these techniques to specific problems by microbiologists can help to guide the development of new techniques and materials by chemists, physicists and engineers.
Abstract
This Review summarizes methods for constructing systems and structures at micron or submicron scales that have applications in microbiology. These tools make it possible to manipulate individual cells and their immediate extracellular environments and have the capability to transform the study of microbial physiology and behaviour. Because of their simplicity, low cost and use in microfabrication, we focus on the application of soft lithographic techniques to the study of microorganisms, and describe several key areas in microbiology in which the development of new microfabricated materials and tools can have a crucial role.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Xia, Y. & Whitesides, G. M. Soft lithography. Angewandte Chemie, International Edition 37, 550–575 (1998). This review provides an overview of the techniques of soft lithography and their application in the fabrication of microstructured and nanostructured materials.
Whitesides, G. M., Ostuni, E., Takayama, S., Jiang, X. & Ingber, D. E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 3, 335–373 (2001). This review introduces several applications of microstructures in mammalian cell biology and provides examples of biological questions that might be studied with these materials
Moller-Jensen, J. & Loewe, J. Increasing complexity of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 17, 75–81 (2005).
Bates, D. & Kleckner, N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121, 899–911 (2005).
Ryan, K. R. & Shapiro, L. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72, 367–394 (2003).
Shapiro, L., McAdams, H. H. & Losick, R. Generating and exploiting polarity in Bacteria. Science 298, 1942–1946 (2002).
Lutkenhaus, J. Dynamic proteins in bacteria. Curr. Opin. Microbiol. 5, 548–552 (2002).
Margolin, W. Bacterial cell division: A moving MinE sweeper boggles the MinD. Curr. Biol. 11, R395–R398 (2001).
Bassler, B. L. & Losick, R. Bacterially speaking. Cell 125, 237–246 (2006).
Belas, R. in Bacteria as Multicellular Organisms (J. A. Shapiro and M. Dworkin) 183–219 (1997).
Shapiro, J. A. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104 (1998).
Chen, C. S., Jiang, X. & Whitesides, G. M. Microengineering the environment of mammalian cells in culture. MRS Bulletin 30, 194–201 (2005).
Jiang, X. & Whitesides, G. M. Engineering microtools in polymers to study cell biology. Eng. Life Sci. 3, 475–480 (2003).
Curtis, T. P., Sloan, W. T. & Scannell, J. W. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA 99, 10494–10499 (2002).
Rappe, M. S. & Giovannoni, S. J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Brehm-Stecher, B. F. & Johnson, E. A. Single-cell microbiology: tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 68, 538 (2004).
Kitano, H. Systems biology: a brief overview. Science 295, 1662–1664 (2002).
Breslauer, D. N., Lee, P. J. & Lee, L. P. Microfluidics-based systems biology. Mol. Biosyst. 2, 97–112 (2006). The authors describe applications of microfluidic structures to the study of systems biology.
Gates, B. D., Xu, Q., Love, J. C., Wolfe, D. B. & Whitesides, G. M. Unconventional nanofabrication. Annu. Rev. Materials Res. 34, 339–372 (2004).
Wolfe, D. B. & Whitesides, G. M. in Nanolithography and Patterning Techniques in Microelectronics (ed. D. Bucknall) 76–119 (Woodhead publishing limited, Cambridge UK, 2005).
Linder, V., Wu, H. K., Jiang, X. Y. & Whitesides, G. M. Rapid prototyping of 2D structures with feature sizes larger than 8 mu m. Anal. Chem. 75, 2522–2527 (2003).
Duffy, D. C., McDonald, J. C., Schueller, O. J. A. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Lee, J. N., Jiang, X., Ryan, D. & Whitesides, G. M. Compatibility of mammalian cells on surfaces of poly(dimethylsiloxane). Langmuir 20, 11684–11691 (2004).
Quist, A. P., Pavlovic, E. & Oscarsson, S. Recent advances in microcontact printing. Anal. Bioanal. Chem. 381, 591–600 (2005).
Delamarche, E. Microcontact printing of proteins. Nanobiotechnol. 31–52 (2004).
Weibel, D. B. et al. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Langmuir 21, 6436–6442 (2005). The authors demonstrate a technique for stamping patterns of bacteria on surfaces that are relevant to the study of chemical and physical interactions between microbes.
Love, J. C., Estroff, L. A., Kriebel, J. K., Nuzzo, R. G. & Whitesides, G. M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chemical Reviews 105, 1103–1169 (2005).
Campbell, C. J., Smoukov, S. K., Bishop, K. J. M. & Grzybowski, B. A. Reactive surface micropatterning by wet stamping. Langmuir 21, 2637–2640 (2005).
Mayer, M., Yang, J., Gitlin, I., Gracias, D. H. & Whitesides, G. M. Micropatterned agarose gels for stamping arrays of proteins and gradients of proteins. Proteomics 4, 2366–2376 (2004).
Whitesides, G. M. The origins and the future of microfluidics. Nature 442, 368–373 (2006). This review is one of several articles in a Nature Insight section that provides an excellent overview of the history and applications of microfluidics.
McDonald, J. C. et al. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21, 27–40 (2000).
Squires, T. M. & Quake, S. R. Microfluidics: fluid physics at the nanoliter scale. Reviews of Modern Physics 77, 977–1026 (2005).
Jeon, N. L. et al. Generation of solution and surface gradients using microfluidic systems. Langmuir 16, 8311–8316 (2000).
Dertinger, S. K. W., Chiu, D. T., Jeon, N. L. & Whitesides, G. M. Generation of gradients having complex shapes using microfluidic networks. Anal. Chem. 79, 1240–1246 (2001).
Jiang, X. Y. et al. A general method for patterning gradients of biomolecules on surfaces using microfluidic networks. Anal. Chem. 77, 2338–2347 (2005).
Unger, M. A., Chou, H. P., Thorsen, T., Scherer, A. & Quake, S. R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000).
Thorsen, T., Maerkl, S. J. & Quake, S. R. Microfluidic large-scale integration. Science 298, 580–584 (2002).
Liang, M. N. et al. Measuring the forces involved in polyvalent adhesion of uropathogenic Escherichia coli to mannose-presenting surfaces. Proc. Natl Acad. Sci. USA 97, 13092–13096 (2000).
Rowan, B., Wheeler, M. A. & Crooks, R. M. Patterning bacteria within hyperbranched polymer film templates. Langmuir 18, 9914–9917 (2002).
St John, P. M. et al. Diffraction-based cell detection using a microcontact printed antibody grating. Anal. Chem. 70, 1108–1111 (1998).
Morhard, F., Pipper, J., Dahint, R. & Grunze, M. Immobilization of antibodies in micropatterns for cell detection by optical diffraction. Sens Actuators B-Chem. 70, 232–242 (2000).
Howell, S. W., Inerowicz, H. D., Regnier, F. E. & Reifenberger, R. Patterned protein microarrays for bacterial detection. Langmuir 19, 436–439 (2003).
Rozhok, S. et al. Methods for fabricating microarrays of motile bacteria. Small 1, 445–451 (2005).
Suh, K. Y., Khademhosseini, A., Yoo, P. J. & Langer, R. Patterning and separating infected bacteria using host–parasite and virus–antibody interactions. Biomed. Microdevices 6, 223–229 (2004).
Heo, J., Thomas, K. J., Seong, G. H. & Crooks, R. M. A microfluidic bioreactor based on hydrogel-entrapped E. coli: Cell viability, lysis, and intracellular enzyme reactions. Anal. Chem. 75, 22–26 (2003).
Beebe, D. J., Mensing, G. A. & Walker, G. M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 4, 261–286 (2002).
DiLuzio, W. R. et al. Escherichia coli swim on the right-hand side. Nature 435, 1271–1274 (2005). The authors use microfluidics to study the interactions of motile cells of bacteria with surfaces. They observe that hydrodynamic interactions between cells and the walls of the channels cause the cells to swim preferentially on one side of the channel.
Mao, H. B., Cremer, P. S. & Manson, M. D. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl Acad. Sci. USA 100, 5449–5454 (2003).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Park, S. et al. Motion to form a quorum. Science 301, 188 (2003).
Park, S. et al. Influence of topology on bacterial social interaction. Proc. Natl Acad. Sci. USA 100, 13910–13915 (2003).
Keymer, J. E., Galajda, P., Muldoon, C., Park, S. & Austin, R. H. Bacterial meta populations in nanofabricated landscapes. Proc. Natl Acad. Sci. USA 103, 17290–17295 (2006). Using microstructured and nanostructured materials the authors studied the interaction and adaptation of cells of bacteria in confined environments.
Takeuchi, S., DiLuzio, W. R., Weibel, D. B. & Whitesides, G. M. Controlling the shape of filamentous cells of Escherichia coli. Nano Letters 5, 1819–1823 (2005). The authors demonstrate an approach for engineering the shape of bacteria by growing filamentous cells in microfabricated compartments with a defined shape.
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004). Using a microfluidic flow cell the authors confined individual cells of bacteria in channels and studied the response of 'persister' cells to treatment with antibiotics.
Novick, A. & Szilard, L. Description of the chemostat. Science 112, 715–716 (1950).
Zhang, Z. et al. Microchemostat-microbial continuous culture in a polymer-based, instrumented microbioreactor. Lab Chip 6, 906–913 (2006).
Balagadde, F. K., You, L. C., Hansen, C. L., Arnold, F. H. & Quake, S. R. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 309, 137–140 (2005).
Groisman, A. et al. A microfluidic chemostat for experiments with bacterial and yeast cells. Nature Methods 2, 685–689 (2005).
Darnton, N., Turner, L., Breuer, K. & Berg, H. C. Moving fluid with bacterial carpets. Biophys. J. 86, 1863–1870 (2004).
Kim, M. J. & Breuer, K. S. Enhanced diffusion due to motile bacteria. Phys. Fluids 16, L78–L81 (2004).
Hiratsuka, Y., Miyata, M. & Uyeda, T. Q. P. Living microtransporter by uni-directional gliding of Mycoplasma along microtracks. Biochem. Biophys. Res. Commun. 331, 318–324 (2005).
Hiratsuka, Y., Miyata, M., Tada, T. & Uyeda, T. Q. P. A microrotary motor powered by bacteria. Proc. Natl Acad. Sci. USA 103, 13618–13623 (2006).
Weibel, D. B. et al. Microoxen: microorganisms to move microscale loads. Proc. Natl Acad. Sci. USA 102, 11963–11967 (2005).
Kim, J. et al. Cell lysis on a microfluidic CD (compact disc). Lab Chip 4, 516–522 (2004).
Enger, J., Goksoer, M., Ramser, K., Hagberg, P. & Hanstorp, D. Optical tweezers applied to a microfluidic system. Lab Chip 4, 196–200 (2004).
Nagamine, K. et al. On-chip transformation of bacteria. Anal. Chem. 77, 4278–4281 (2005).
Kricka, L. J. & Wilding, P. Microchip PCR. Anal. Bioanal. Chem. 377, 820–825 (2003).
Roper, M. G., Easley, C. J. & Landers, J. P. Advances in polymerase chain reaction on microfluidic chips. Anal. Chem. 77, 3887–3893 (2005).
Cady, N. C., Stelick, S., Kunnavakkam, M. V. & Batt, C. A. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens Actuators B-Chem. 107, 332–341 (2005).
Hong, J. W., Studer, V., Hang, G., Anderson, W. F. & Quake, S. R. A nanoliter-scale nucleic acid processor with parallel architecture. Nature Biotechnol. 22, 435–439 (2004).
Hong, J. W., Chen, Y., Anderson, W. F. & Quake, S. R. Molecular biology on a microfluidic chip. J. Phys. Condens. Matter 18, S691–S701 (2006).
Ottesen, E. A., Hong, J. W., Quake, S. R. & Leadbetter, J. R. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467 (2006).
Ostuni, E., Kane, R., Chen, C., Ingber, D. & Whitesides, G. Patterning mammalian cells using elastomeric membranes. Langmuir 16, 7811–7819 (2000).
Acknowledgements
This work was supported by grants from the National Institutes of Health and the Defense Advanced Research Projects Agency. We thank Rich Losick for reading the manuscript and providing insightful comments.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
George M. Whitesides' homepage
Harvard University Soft Lithography Foundry
Glossary
- Microtechnology
-
The fabrication and application of materials, structures and systems with micron or submicron-scale features.
- Microenvironment
-
The region sensed by a cell. The dimensions of this region are usually set by molecular contact, mass transport and diffusion, and range from a few nanometers to perhaps a millimeter.
- Soft lithography
-
A set of techniques that makes microstructures by printing, moulding and embossing using a patterned, elastomeric stamp or mould, and/or a polymeric substrate.
- Soft material
-
A material (especially a polymer) that is pliable, compressible or elastic.
- Public foundry
-
A facility for the fabrication of microstructured materials and systems.
- Elastomeric polymer
-
A soft, compliant, rubber-like polymer.
- Mask
-
Typically a transparent substrate with a pattern on its surface defined in an opaque material (chrome metal or ink) used in photolithography.
- PDMS
-
Poly(dimethylsiloxane). An elastomeric silicone polymer that is commercially available and has properties that make it well suited to applications in microbiology.
- Embossed structure
-
A structure that is moulded in a surface in relief.
- Bas-relief structure
-
A structure that projects away from a surface.
- Bas-relief master
-
Refers to the master copy and, in soft lithography, consists of patterns of a photoreactive polymer on the surface of a silicon wafer or glass slide.
- Photolithography
-
A process used to transfer a pattern from a mask onto a thin film of photosensitive polymer (photoresist) and then onto the surface of a substrate. Photolithography is commonly used in semiconductor fabrication to fabricate integrated circuits.
- CAD tool
-
Computer aided design. A software program used by engineers and designers for drafting two- and three-dimensional structures.
- Photoresist
-
A photoreactive polymer that undergoes chemical changes that lead to changes in physical properties (such as solubility) after exposure to ultraviolet light.
- Microfluidic system
-
A set of channels that have micron-scale dimensions (typically between 5–500 μm), and are used to manipulate fluids.
- Spin coating
-
A process for depositing uniform layers of polymer on a substrate. Rotating the substrate at a high speed spreads the material uniformly over the surface. The viscosity of the material and the rotational velocity of the substrate control the thickness of the layer of material; surface tension flattens the surface of the spun film.
- SAMs
-
Self assembled monolayers. Monolayer structures formed by the spontaneous self-assembly of alkanethiols on metal surfaces. In SAMs, the thiol groups are bonded covalently to the metal surface, and the non-covalent, intermolecular packing of the alkane chains causes the molecules to arrange into an ordered, two-dimensional crystal or liquid crystal.
- Hydrogel
-
A low-density, crosslinked polymer network containing a high-volume fraction of water.
Rights and permissions
About this article
Cite this article
Weibel, D., DiLuzio, W. & Whitesides, G. Microfabrication meets microbiology. Nat Rev Microbiol 5, 209–218 (2007). https://doi.org/10.1038/nrmicro1616
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1616
This article is cited by
-
Microfluidics for adaptation of microorganisms to stress: design and application
Applied Microbiology and Biotechnology (2024)
-
Single-cell pathogen diagnostics for combating antibiotic resistance
Nature Reviews Methods Primers (2023)
-
A ‘print–pause–print’ protocol for 3D printing microfluidics using multimaterial stereolithography
Nature Protocols (2023)
-
Microbial Fuel Cell–Based Biosensors and Applications
Applied Biochemistry and Biotechnology (2023)
-
Cell phone microscopy enabled low-cost manufacturable colorimetric urine glucose test
Biomedical Microdevices (2023)