Key Points
-
Virus Particle Explorer (VIPER: http://mmtsb.scripps.edu/viper) is a web-based catalogue of structural information describing icosahedral virus particles.
-
Virus atomic coordinates obtained from crystallographic methods conform to a specific standard icosahedral orientation in VIPER, allowing for straightforward global comparisons and en-masse calculation of derived information. Each viral capsid structure is represented pictorially and with an extensive computational analysis highlighting inter-subunit residue contacts, binding energies, quasi-equivalence and proposed assembly pathways.
-
VIPER provides a series of web-based tools that further allow users to visualize and explore the data provided: 'Oligomer Generator' to obtain coordinates for the entire viral capsid or a specified portion of it; 'Icosahedral Server', an educational tool to understand the geometric principles for generating icosahedral quasi-equivalent surface lattices; 'Map a Residue' to display a user-specified set of residues on the subunit of a chosen virus structure; 'Visual VIPER' to visually compare properties such as topology, subunit structure, subunit organization, crystal contacts and so on among a chosen set of virus structures.
-
VIPER currently includes the electron density available for low–medium-resolution virus structures obtained by cryo-electron microscopy (EM) techniques that will complement the existing high-resolution crystal structures in the database.
-
The VIPER database with visualization and analysis for virus structural data obtained from two different experimental techniques will be a structural resource for virologists, microbiologists, virus crystallographers and EM researchers.
Abstract
Virus structures are megadalton nucleoprotein complexes with an exceptional variety of protein–protein and protein–nucleic-acid interactions. Three-dimensional crystal structures of over 70 virus capsids, from more than 20 families and 30 different genera of viruses, have been solved to near-atomic resolution. The enormous amount of information contained in these structures is difficult to access, even for scientists trained in structural biology. Virus Particle Explorer (VIPER) is a web-based catalogue of structural information that describes the icosahedral virus particles. In addition to high-resolution crystal structures, VIPER has expanded to include virus structures obtained by cryo-electron microscopy (EM) techniques. The VIPER database is a powerful resource for virologists, microbiologists, virus crystallographers and EM researchers. This review describes how to use VIPER, using several examples to show the power of this resource for research and educational purposes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Reddy, V. S. et al. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 75, 11943–11947 (2001).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 25, 31–36 (1997).
Brooks, B. R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4, 187–217 (1983).
Eisenberg, D. & McLachlan, A. D. Solvation energy in protein folding and binding. Nature 319, 199–203 (1986).
Reddy, V. S. et al. Energetics of quasiequivalence: computational analysis of protein–protein interactions in icosahedral viruses. Biophys. J. 74: 546–558 (1998). This paper provides information on identifying unique protein–protein interfaces and calculation of association energies for these interfaces to explore possible assembly pathways for various virus structures.
Pettersen, E. F. et al. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Walther, D. WebMol — a Java based PDB viewer. Trends Biochem. Sci. 22, 274–275 (1997).
Hogue, C. W. V. Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem. Sci., 22, 314–316 (1997).
Natarajan, P. & Johnson, J. E. Molecular packing in virus crystals: geometry, chemistry, and biology. J. Struct. Biol. 121, 295–305 (1998). This paper presents a method to extract the details of inter-particle protein–protein interactions in virus crystals and shows a correlation between the residues buried in crystal contacts and the residues involved in biological processes such as receptor or antibody binding.
Caspar, D. L. D. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27, 1–24 (1962).
Damodaran, K. V., Reddy, V. S., Johnson, J. E. & Brooks, C. L. 3rd. A general method to quantify quasi-equivalence in icosahedral viruses. J. Mol. Biol. 324, 723–737 (2002).
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
DeLano, W. L. The PyMOL Molecular Graphics System [online] <http://www.pymol.org> (2002).
Acknowledgements
The development and support of VIPER is directed through the Center for Multiscale Modeling Tools for Structural Biology and a NIH-funded Research Resource.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez
FURTHER INFORMATION
Glossary
- ASYMMETRIC UNIT
-
The asymmetric unit of an icosahedral virus structure is defined as the smallest part of the structure from which the complete structure of the virus can be built using a specific set of 60 rotational matrices that describe the 5:3:2 symmetry of the virus particle.
- CAPSOMERES
-
The obvious surface features of the virus particle, which are observable in an electron-microscopy-reconstructed density.
- OLIGOMER
-
The representation of the protein subunit on the viral capsid as dimer, trimer, pentamer, hexamer and so on.
- T (TRIANGULATION) NUMBER
-
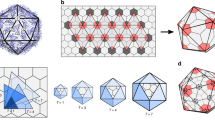
The theoretical basis for the structure of isometric viruses was described by Caspar and Klug with their concept of identical elements in quasi-equivalent environments. They defined all possible polyhedra in terms of structure units. The icosahedron itself has 20 equilateral triangular facets, and therefore 20 T structure units, in which T is the triangulation number given by the rule T = Pf, in which P can be any number of the series 1, 3, 7, 13, 19, 21, 31 and so on (= h2 + hk +k2, for all pairs of integers, h and k having no common factor) and f is any integer.
- ICOSAHEDRON
-
A polyhedron comprising 20 equilateral triangular facets and 12 vertices that has rotational symmetry described as 5:3:2 symmetry. There are six 5-fold axes of symmetry passing through the vertices, ten 3-fold axes extending through each face and fifteen 2-fold axes passing through the edges of an icosahedron. The VIPER database only holds data for icosahedral virus structures.
- C-α TRACING
-
A simplified representation of the tertiary structure of the subunit, in which a C-α atom represents each residue and the subsequent C-α atoms are connected by a line.
- CAGE
-
The geometric representation of the icosahedral symmetry of a virus particle.
- VRML
-
Virtual reality modelling language used in three-dimensional viewing of the tertiary structure of subunits in the VIPER tool 'Map a Residue'.
Rights and permissions
About this article
Cite this article
Natarajan, P., Lander, G., Shepherd, C. et al. Exploring icosahedral virus structures with VIPER. Nat Rev Microbiol 3, 809–817 (2005). https://doi.org/10.1038/nrmicro1283
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1283
This article is cited by
-
Structure of icosahedral quasicrystals within the multiple-cell approach
Structural Chemistry (2020)
-
Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM
Nature Communications (2015)
-
Physical virology
Nature Physics (2010)