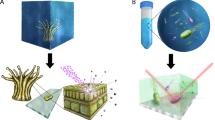
Abstract
Stable isotope probing (SIP) is a technique that is used to identify the microorganisms in environmental samples that use a particular growth substrate. The method relies on the incorporation of a substrate that is highly enriched in a stable isotope, such as 13C, and the identification of active microorganisms by the selective recovery and analysis of isotope-enriched cellular components. DNA and rRNA are the most informative taxonomic biomarkers and 13C-labelled molecules can be purified from unlabelled nucleic acid by density-gradient centrifugation. The future holds great promise for SIP, particularly when combined with other emerging technologies such as microarrays and metagenomics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rappé, M. S. & Giovannoni, S. J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003).
Zengler, K. et al. Cultivating the uncultured. Proc. Natl Acad. Sci. USA 99, 15681–15686 (2002).
Cole, J. R. et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33, 294–296 (2005).
Schloss, P. D. & Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68, 686–691 (2004).
Ouverney, C. C. & Fuhrman, J. A. Combined microautoradiography–16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65, 1746–1752 (1999).
Lee, N. et al. Combination of fluorescent in situ hybridization and microautoradiography — a new tool for structure–function analyses in microbial ecology. Appl. Environ. Microbiol. 65, 1289–1297 (1999).
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K. H. & Wagner, M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284 (2001).
Adamczyk, J. et al. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69, 6875–6887 (2003).
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D. & DeLong, E. F. Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487 (2001).
Boetius, A. et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626 (2000).
Boschker, H. T. S. et al. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392, 801–805 (1998).
Radajewski, S., Ineson, P., Parekh, N. R. & Murrell, J. C. Stable-isotope probing as a tool in microbial ecology. Nature 403, 646–649 (2000).
Meselson, M. & Stahl, F. W. The replication of DNA in Escherichia coli. Proc. Natl Acad. Sci. USA 44, 671–682 (1958).
Schildkraut, C. L. in Methods in Enzymology (eds Grossman, L. & Moldave, K.) 695–699 (Academic Press, New York, 1967).
Radajewski, S. & Murrell, J. C. Stable isotope probing for detection of methanotrophs after enrichment with 13CH4 . Methods Mol. Biol. 179, 149–157 (2002).
Bodrossy, L. et al. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5, 566–582 (2003).
Dedysh, S. N. et al. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282, 281–284 (1998).
Morris, S. A., Radajewski, S., Willison, T. W. & Murrell, J. C. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68, 1446–1453 (2002).
Radajewski, S. et al. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148, 2331–2342 (2002).
Hutchens, E., Radajewski, S., Dumont, M. G., McDonald, I. R. & Murrell, J. C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6, 111–120 (2004).
Lin, J. -L. et al. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6, 1049–1060 (2004).
Whitby, C. B. et al. 13C incorporation into DNA as a means of identifying the active components of ammonia-oxidizer populations. Lett. Appl. Microbiol. 32, 398–401 (2001).
Miller, L. G. et al. Degradation of methyl bromide and methyl chloride in soil microcosms: use of stable C isotope fractionation and stable isotope probing to identify reactions and the responsible microorganisms. Geochim. Cosmochim. Acta 68, 3271–3283 (2004).
McDonald, I. R. et al. A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ. Microbiol. 4, 193–203 (2002).
Ginige, M. P. et al. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization–microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70, 588–596 (2004).
Padmanabhan, P. et al. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl. Environ. Microbiol. 69, 1614–1622 (2003).
Jeon, C. O. et al. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl Acad. Sci. USA 100, 13591–13596 (2003).
Wackett, L. P. Stable isotope probing in biodegradation research. Trends Biotechnol. 22, 153–154 (2004).
Manefield, M., Whiteley, A. S. & Bailey, M. J. What can stable isotope probing do for bioremediation? Inter. Biodeter. Biodegradation 54, 163–166 (2004).
Lueders, T., Pommerenke, B. & Friedrich, M. W. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70, 5778–5786 (2004).
Manefield, M., Whiteley, A. S., Griffiths, R. I. & Bailey, M. J. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68, 5367–5373 (2002).
Manefield, M., Whiteley, A. S., Ostle, N., Ineson, P. & Bailey, M. J. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16, 2179–2183 (2002).
Mahmood, S., Paton, G. I. & Prosser, J. I. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. (in the press).
Lueders, T., Manefield, M. & Friedrich, M. W. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78 (2004).
Lueders, T., Wagner, B., Claus, P. & Friedrich, M. W. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6, 60–72 (2004).
Singh, B. K., Millard, P., Whiteley, A. S. & Murrell, J. C. Unravelling rhizosphere–microbial interactions: opportunities and limitations. Trends Microbiol. 12, 386–393 (2004).
Griffiths, R. I. et al. 13CO2 pulse labelling of plants in tandem with stable isotope probing: methodological considerations for examining microbial function in the rhizosphere. J. Microbiol. Methods 58, 119–129 (2004).
Rangel-Castro, J. I. et al. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ. Microbiol. (in the press).
Schloss, P. D. & Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14, 303–310 (2003).
Handelsman, J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685 (2004).
Rondon, M. R. et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547 (2000).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso sea. Science 304, 66–74 (2004).
Tyson, G. W. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004).
Radajewski, S., McDonald, I. R. & Murrell, J. C. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14, 296–302 (2003).
Bull, I. D., Parekh, N. R., Hall, G. H., Ineson, P. & Evershed, R. P. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature 405, 175–178 (2000).
Acknowledgements
M.G.D. received financial support during his Ph.D. from the Fonds de Recherche sur la Nature et les Technologies (Quebec, Canada). J.C.M. gratefully acknowledges support from the Natural Environment Research Council, the Biotechnology and Biological Sciences Research Council and the European Union for funding work in his laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- CONSORTIUM
-
Physical association between cells of two or more types of microorganisms. Such an association might be advantageous to at least one of the microorganisms.
- SYNTROPHIC ASSOCIATION
-
An association in which the growth of one organism depends on, or is improved by, growth factors or substrates provided (or in some cases removed) by another organism growing nearby.
Rights and permissions
About this article
Cite this article
Dumont, M., Murrell, J. Stable isotope probing — linking microbial identity to function. Nat Rev Microbiol 3, 499–504 (2005). https://doi.org/10.1038/nrmicro1162
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1162
This article is cited by
-
The sustainable approach of microbial bioremediation of arsenic: an updated overview
International Journal of Environmental Science and Technology (2024)
-
Potential of stable isotope analysis to deduce anaerobic biodegradation of ethyl tert-butyl ether (ETBE) and tert-butyl alcohol (TBA) in groundwater: a review
Environmental Science and Pollution Research (2024)
-
Microbial controls over soil priming effects under chronic nitrogen and phosphorus additions in subtropical forests
The ISME Journal (2023)
-
Differences among active toluene-degrading microbial communities in farmland soils with different levels of heavy metal pollution
Biodegradation (2023)
-
Functional and molecular approaches for studying and controlling microbial communities in anaerobic digestion of organic waste: a review
Reviews in Environmental Science and Bio/Technology (2023)