Key Points
-
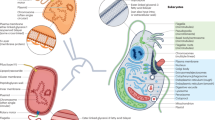
Horizontal gene transfer (HGT) is essential for genome evolution across the tree of life and has an important role in archaeal speciation, adaptation, and maintenance of diversity.
-
Many of the horizontally acquired genes in archaea are involved in metabolism and cell envelope biogenesis and therefore likely provided a selective advantage for adaptation to new environments.
-
The transfer of DNA can also function in DNA repair by providing an intact template for homologous recombination, as has been shown for Sulfolobus spp.
-
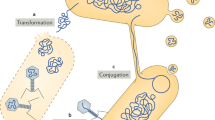
Classic bacterial DNA transfer mechanisms, such as transformation, conjugation and transduction, have been identified in certain archaeal species. In addition, DNA exchange through vesicles, through cell fusion and by the recently discovered archaea-specific crenarchaeal exchange of DNA (Ced) system has been observed.
-
Several barriers prevent the transfer and incorporation of foreign DNA in archaeal species; these include physical environmental barriers, the surface layer (S-layer), CRISPR–Cas systems, restriction–modification systems and toxin–antitoxin systems.
-
Newly identified and sequenced species will enable us to uncover more HGT events among and between archaea, bacteria and eukaryotes.
-
With an increasing number of genetically tractable archaeal species, we will be able to elucidate the processes that underlie gene flow in archaea.
Abstract
Archaea are diverse, ecologically important, single-celled microorganisms. They have unique functions and features, such as methanogenesis and the composition of their cell envelope, although many characteristics are shared with the other domains of life, either through ancestry or through promiscuous horizontal gene transfer. The exchange of genetic material is a major driving force for genome evolution across the tree of life and has a role in archaeal speciation, adaptation and maintenance of diversity. In this Review, we discuss our current knowledge of archaeal mechanisms of DNA transfer and highlight the role of gene transfer in archaeal evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Cavicchioli, R. Cold-adapted archaea. Nat. Rev. Microbiol. 4, 331–343 (2006).
Chen, X.-P., Zhu, Y.-G., Xia, Y., Shen, J.-P. & He, J.-Z. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10, 1978–1987 (2008).
Offre, P., Spang, A. & Schleper, C. Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457 (2013).
Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8, 731–741 (2010).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Guy, L. & Ettema, T. J. G. The archaeal 'TACK' superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587 (2011).
Williams, T. A., Foster, P. G., Nye, T. M. W., Cox, C. J. & Embley, T. M. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc. Biol. Sci. 279, 4870–4879 (2012).
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579 (1990).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Nelson-Sathi, S. et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl Acad. Sci. USA 109, 20537–20542 (2012). This paper suggests that a single gene transfer event transformed a methanogen in the last haloarchaeal common ancestor.
Cadillo-Quiroz, H. et al. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 10, e1001265 (2012).
Whitaker, R. J., Grogan, D. W. & Taylor, J. W. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol. Biol. Evol. 22, 2354–2361 (2005).
Papke, R. T., Koenig, J. E., Rodríguez-Valera, F. & Doolittle, W. F. Frequent recombination in a saltern population of Halorubrum. Science 306, 1928–1929 (2004).
Nelson-Sathi, S. et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80 (2015). This study provides evidence for high bacteria-to-archaea gene transfer at the origin of deeply branching archaeal taxa.
Worrell, V. E., Nagle, D. P., McCarthy, D. & Eisenbraun, A. Genetic transformation system in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 170, 653–656 (1988).
Held, N. L., Herrera, A., Cadillo-Quiroz, H. & Whitaker, R. J. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS ONE 5, e12988 (2010).
Schleper, C., Holz, I., Janekovic, D., Murphy, J. & Zillig, W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177, 4417–4426 (1995). This study describes the first observation of an archaeal conjugative plasmid.
Naor, A., Lapierre, P., Mevarech, M., Papke, R. T. & Gophna, U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr. Biol. 22, 1444–1448 (2012).
Naor, A. & Gophna, U. Cell fusion and hybrids in Archaea: prospects for genome shuffling and accelerated strain development for biotechnology. Bioengineered 4, 9–8 (2013).
van Wolferen, M., Wagner, A., van der Does, C. & Albers, S.-V. The archaeal Ced system imports DNA. Proc. Natl Acad. Sci. USA 113, 2496–2501 (2016). This study, for the first time, describes proteins that are involved in DNA transport machinery in archaea.
Brochier-Armanet, C. et al. Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J. 5, 1291–1302 (2011).
Deschamps, P., Zivanovic, Y., Moreira, D., Rodriguez-Valera, F. & López-García, P. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol. Evol. 6, 1549–1563 (2014).
López-García, P., Zivanovic, Y., Deschamps, P. & Moreira, D. Bacterial gene import and mesophilic adaptation in archaea. Nat. Rev. Microbiol. 13, 447–456 (2015).
Allen, M. A. et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 3, 1012–1035 (2009).
Deppenmeier, U. et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461 (2002).
Raymann, K., Forterre, P., Brochier-Armanet, C. & Gribaldo, S. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol. Evol. 6, 192–212 (2014).
Wang, J. C. DNA topoisomerases. Annu. Rev. Biochem. 65, 635–692 (1996).
López-García, P. DNA supercoiling and temperature adaptation: a clue to early diversification of life? J. Mol. Evol. 49, 439–452 (1999).
Groussin, M. & Gouy, M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol. Biol. Evol. 28, 2661–2674 (2011).
Williams, D., Gogarten, J. P. & Papke, R. T. Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol. Evol. 4, 1223–1244 (2012).
DeMaere, M. Z. et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl Acad. Sci. USA 110, 16939–16944 (2013).
Papke, R. T. et al. Horizontal gene transfer, dispersal and haloarchaeal speciation. Life (Basel) 5, 1405–1426 (2015).
Held, N. L., Herrera, A. & Whitaker, R. J. Reassortment of CRISPR repeat-spacer loci in Sulfolobus islandicus. Environ. Microbiol. 11, 3065–3076 (2013).
Krause, D. J. & Whitaker, R. J. Inferring speciation processes from patterns of natural variation in microbial genomes. Syst. Biol. 64, 926–935 (2015).
Reno, M. L., Held, N. L., Fields, C. J., Burke, P. V. & Whitaker, R. J. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl Acad. Sci. USA 106, 8605–8610 (2009).
Brock, T. D., Brock, K. M., Belly, R. T. & Weiss, R. L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68 (1972).
Naor, A., Lazary, R., Barzel, A., Papke, R. T. & Gophna, U. In vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii. PLoS ONE 6, e15833 (2011).
Grogan, D. W. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178, 3207–3211 (1996).
Ghane, F. & Grogan, D. W. Chromosomal marker exchange in the thermophilic archaeon Sulfolobus acidocaldarius: physiological and cellular aspects. Microbiology 144, 1649–1657 (1998).
Fröls, S. et al. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70, 938–952 (2008).
Ajon, M. et al. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817 (2011).
Chen, I. & Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 (2004).
Salmond, G. P. C. & Fineran, P. C. A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786 (2015).
Guglielmini, J. et al. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715–5727 (2014).
Gaudin, M. et al. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Env. Microbiol. Rep. 5, 109–116 (2013). This paper describes, for the first time, the transfer of archaeal plasmids through vesicles.
Bertani, G. & Baresi, L. Genetic transformation in the methanogen Methanococcus voltae PS. J. Bacteriol. 169, 2730–2738 (1987).
Sato, T., Fukui, T., Atomi, H. & Imanaka, T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220 (2003).
Waege, I., Schmid, G., Thumann, S., Thomm, M. & Hausner, W. Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Env. Microbiol. 76, 3308–3313 (2010).
Grogan, D. W. & Stengel, K. R. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli /λRed and Sulfolobus acidocaldarius recombination systems. Mol. Microbiol. 69, 1255–1265 (2008).
Patel, G. B., Nash, J. H., Agnew, B. J. & Sprott, G. D. Natural and electroporation-mediated transformation of Methanococcus voltae protoplasts. Appl. Environ. Microbiol. 60, 903–907 (1994).
Sato, T., Fukui, T., Atomi, H. & Imanaka, T. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Env. Microbiol. 71, 3889–3899 (2005).
Lipscomb, G. L. et al. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Env. Microbiol. 77, 2232–2238 (2011).
Averhoff, B. Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33, 611–626 (2009).
Claverys, J.-P., Martin, B. & Polard, P. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33, 643–656 (2009).
Gould, S. B. et al. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol. 24, 525–534 (2016).
Soler, N., Marguet, E., Verbavatz, J.-M. & Forterre, P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 159, 390–399 (2008).
Choi, D. H. et al. Extracellular vesicles of the hyperthermophilic archaeon Thermococcus onnurineus NA1 T. Appl. Environ. Microbiol. 81, 4591–4599 (2015).
Soler, N., Gaudin, M., Marguet, E. & Forterre, P. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem. Soc. Trans. 39, 36–44 (2011).
Gaudin, M. et al. Extracellular membrane vesicles harbouring viral genomes. Environ. Microbiol. 16, 1167–1175 (2014).
Santangelo, T. J., Cubonová, L. & Reeve, J. N. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Env. Microbiol. 74, 3099–3104 (2008).
Shetty, A., Chen, S., Tocheva, E. I., Jensen, G. J. & Hickey, W. J. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS ONE 6, e20725 (2011).
Marguet, E. et al. Membrane vesicles, nanopods and/or nanotubes produced by hyperthermophilic archaea of the genus Thermococcus. Biochem. Soc. Trans. 41, 436–442 (2013).
Ellen, A. F. et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13, 67–79 (2009).
Prangishvili, D. et al. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 182, 2985–2988 (2000).
Mashburn-Warren, L. M. & Whiteley, M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61, 839–846 (2006).
Kulp, A. & Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 (2010).
Deatherage, B. L. & Cookson, B. T. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957 (2012).
Nieuwland, R. & Sturk, A. Why do cells release vesicles? Thromb. Res. 125 (Suppl.), S49–S51 (2010).
Rice, G. et al. Viruses from extreme thermal environments. Proc. Natl Acad. Sci. USA 98, 13341–13345 (2001).
Witte, A. et al. Characterization of Natronobacterium magadii phage ΦCh1, a unique archaeal phage containing DNA and RNA. Mol. Microbiol. 23, 603–616 (1997).
Liu, Y. et al. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Mol. Microbiol. 98, 1002–1020 (2015).
Krupovic, M., Spang, A., Gribaldo, S., Forterre, P. & Schleper, C. A thaumarchaeal provirus testifies for an ancient association of tailed viruses with archaea. Biochem. Soc. Trans. 39, 82–88 (2011).
Schleper, C., Kubo, K. & Zillig, W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl Acad. Sci. USA 89, 7645–7649 (1992).
Quemin, E. R. J. et al. Eukaryotic-like virus budding in Archaea. mBio 7, e01439-16 (2016).
Arnold, H. P. et al. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 34, 217–226 (1999).
Wang, Y. et al. A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363, 124–133 (2007).
Iro, M. et al. The lysogenic region of virus φCh1: identification of a repressor-operator system and determination of its activity in halophilic Archaea. Extremophiles 11, 383–396 (2007).
Mei, Y. et al. Induction and preliminary characterization of a novel halophage SNJ1 from lysogenic Natrinema sp. F5. Can. J. Microbiol. 53, 1106–1110 (2007).
Schnabel, H. et al. Halobacterium halobium phage øH. EMBO J. 1, 87–92 (1982).
Zhang, Z. et al. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology 434, 233–241 (2012).
Prangishvili, D., Garrett, R. A. & Koonin, E. V. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117, 52–67 (2006).
Pina, M., Bize, A., Forterre, P. & Prangishvili, D. The archeoviruses. FEMS Microbiol. Rev. 35, 1035–1054 (2011).
Iranzo, J., Koonin, E. V., Prangishvili, D. & Krupovic, M. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J. Virol. 90, 11043–11055 (2016).
Bertani, G. Transduction-like gene transfer in the methanogen Methanococcus voltae. J. Bacteriol. 181, 2992–3002 (1999).
Eiserling, F., Bertani, G., Pushkin, A. & Gingery, M. Bacteriophage-like particles associated with the gene transfer agent of Methanococcus voltae PS. J. Gen. Virol. 80, 3305–3308 (1999).
Lang, A. S., Zhaxybayeva, O. & Beatty, J. T. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482 (2012).
Spang, A. et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14, 3122–3145 (2012).
Redder, P. & Garrett, R. A. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188, 4198–4206 (2006).
She, Q., Peng, X., Zillig, W. & Garrett, R. A. Gene capture in archaeal chromosomes. Nature 409, 478 (2001).
Gudbergsdottir, S. et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79, 35–49 (2011).
Aagaard, C., Dalgaard, J. Z. & Garrett, R. A. Intercellular mobility and homing of an archaeal rDNA intron confers a selective advantage over intron- cells of Sulfolobus acidocaldarius. Proc. Natl Acad. Sci. USA 92, 12285–12289 (1995).
Cavalli, L. L., Lederberg, J. & Lederberg, E. M. An infective factor controlling sex compatibility in Bacterium coli. J. Gen. Microbiol. 8, 89–103 (1953).
Alvarez-Martinez, C. E. & Christie, P. J. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808 (2009).
Cruz, F. De et al. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95 (2014).
Christie, P. J., Whitaker, N. & González-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578–1591 (2014).
Chandran Darbari, V. & Waksman, G. Structural biology of bacterial type IV secretion systems. Annu. Rev. Biochem. 84, 603–629 (2015).
Ilangovan, A., Connery, S. & Waksman, G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 23, 301–310 (2015).
Lacroix, B. & Citovsky, V. Transfer of DNA from bacteria to eukaryotes. mBio 7, e00863-16 (2016).
Prangishvili, D. et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 40, 190–202 (1998).
Stedman, K. M. et al. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J. Bacteriol. 182, 7014–7020 (2000).
Greve, B., Jensen, S., Brugger, K., Zillig, W. & Garrett, R. A. Genomic comparison archaeal conjugative plasmids from Sulfolobus. Archaea 1, 231–239 (2004).
Erauso, G., Stedman, K. M., van den Werken, H. J. G., Zillig, W. & van der Oost, J. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology 152, 1951–1968 (2006).
Basta, T., Smyth, J., Forterre, P., Prangishvili, D. & Peng, X. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 71, 23–34 (2009).
She, Q., Shen, B. & Chen, L. Archaeal integrases and mechanisms of gene capture. Biochem. Soc. Trans. 32, 222–226 (2004).
She, Q. et al. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2, 417–425 (1998).
Wang, H., Peng, N., Shah, S. A., Huang, L. & She, Q. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 79, 117–152 (2015).
Erdmann, S. & Garrett, R. A. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85, 1044–1056 (2012).
Garrett, R. et al. CRISPR–Cas adaptive immune systems of the sulfolobales: unravelling their complexity and diversity. Life 5, 783–817 (2015).
Liu, G., She, Q. & Garrett, R. A. Diverse CRISPR–Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res. 44, 4233–4242 (2016).
Schumacher, M. A. et al. Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science 349, 1120–1124 (2015).
Mevarech, M. & Werczberger, R. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162, 461–462 (1985). This study is the first to show gene transfer in archaea using auxotrophic mutants of H. volcanii.
Fröls, S., Dyall-Smith, M. & Pfeifer, F. Biofilm formation by haloarchaea. Environ. Microbiol. 14, 3159–3174 (2012).
Chimileski, S., Franklin, M. J. & Papke, R. T. Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 12, 65 (2014).
Rosenshine, I., Tchelet, R. & Mevarech, M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245, 1387–1389 (1989).
Fröls, S. et al. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189, 8708–8718 (2007).
Götz, D. et al. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8, R220 (2007).
Fröls, S., White, M. F. & Schleper, C. Reactions to UV damage in the model archaeon Sulfolobus solfataricus. Biochem. Soc. Trans. 37, 36–41 (2009).
Koerdt, A., Gödeke, J., Berger, J., Thormann, K. M. & Albers, S.-V. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 5, e14104 (2010).
Beijersbergen, A., Smith, S. J. & Hooykaas, P. J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32, 212–218 (1994).
Whitaker, R. J., Grogan, D. W. & Taylor, J. W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978 (2003). This paper describes the role and importance of geographical barriers in the evolution of hyperthermophilic archaea.
Bauer, T., Hammes, W. P., Haase, N. U. & Hertel, C. Effect of food components and processing parameters on DNA degradation in food. Environ. Biosafety Res. 3, 215–223 (2004).
Tan, Z.-J. & Chen, S.-J. Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length. Biophys. J. 90, 1175–1190 (2006).
Achtman, M., Kennedy, N. & Skurray, R. Cell–cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc. Natl Acad. Sci. USA 74, 5104–5108 (1977).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 (2010).
Rath, D., Amlinger, L., Rath, A. & Lundgren, M. The CRISPR–Cas immune system: biology, mechanisms and applications. Biochimie 117, 119–128 (2015).
Mruk, I. & Kobayashi, I. To be or not to be: regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 42, 70–86 (2014).
Loenen, W. A. M., Dryden, D. T. F., Raleigh, E. A., Wilson, G. G. & Murray, N. E. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 42, 3–19 (2014).
Oliveira, P. H., Touchon, M. & Rocha, E. P. C. The interplay of restriction–modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 42, 10618–10631 (2014).
Yamaguchi, Y., Park, J.-H. & Inouye, M. Toxin–antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 (2011).
Hazan, R. & Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272, 227–234 (2004).
Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. & Hazan, R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2, e135 (2006).
León-Sobrino, C., Kot, W. P. & Garrett, R. A. Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 99, 719–728 (2016).
Shah, S. A. & Garrett, R. A. in Prokaryotic Toxin–Antitoxins (ed. Gerders, K.) 225–238 (Springer Berlin Heidelberg, 2013).
Zawadzki, P., Roberts, M. S. & Cohan, F. M. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140, 917–932 (1995).
Majewski, J. & Cohan, F. M. Adapt globally, act locally: the effect of selective sweeps on bacterial sequence diversity. Genetics 152, 1459–1474 (1999).
Wolf, Y. I., Rogozin, I. B., Kondrashov, A. S. & Koonin, E. V. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 11, 356–372 (2001).
Majewski, J., Zawadzki, P., Pickerill, P., Cohan, F. M. & Dowson, C. G. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182, 1016–1023 (2000).
Datta, A., Adjiri, A., New, L., Crouse, G. F. & Jinks Robertson, S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 1085–1093 (1996).
Eppley, J. M., Tyson, G. W., Getz, W. M. & Banfield, J. F. Genetic exchange across a species boundary in the archaeal genus Ferroplasma. Genetics 177, 407–416 (2007).
Krause, D. J., Didelot, X., Cadillo-Quiroz, H. & Whitaker, R. J. Recombination shapes genome architecture in an organism from the archaeal domain. Genome Biol. Evol. 6, 170–178 (2014).
Ambur, O.-H., Engelstadter, J., Johnsen, P., Miller, E. & Rozen, D. Steady at the wheel: conservative sex and the benefits of bacterial transformation. Phil. Trans. R. Soc. B. Biol. Sci. 371, 20150528 (2016).
Rocha, E. P. C. Using sex to cure the genome. PLoS Biol. 14, 1–7 (2016).
Mell, J. C. & Redfield, R. J. Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483 (2014).
Johnston, C., Martin, B., Fichant, G., Polard, P. & Claverys, J.-P. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014).
Acknowledgements
R.J.W. and D.J.K. acknowledge funding support from the US National Science Foundaion (NSF; grant DEB #1355171) and NASA Exobiology and Evolutionary Biology (grant NNX09AM92G). A.W., M.v.W. and J.H. were supported by a European Research Council (ERC) starting grant (grant ARCHAELLUM, 311523). J.H. received further support from the CRC746 (DFG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- TACK superphylum
-
A recently proposed superphylum that comprises the Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota phyla.
- Asgard superphylum
-
A recently described superphylum that includes the proposed Lokiarchaeota, Odinarchaeota, Thorarchaeota and Heimdallarchaeota phyla. Asgardarchaeota encode many proteins that were previously suspected to be specific for eukaryotes.
- DPANN superphylum
-
A proposed monophyletic and deep-branching group of mainly hyperthermophilic archaea that includes the Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaeota phyla.
- Heterotrophs
-
Organisms that produce complex organic compounds from organic carbon.
- Autotrophs
-
Organisms that produce complex organic compounds from inorganic material using light energy (photosynthesis) or chemical energy (chemosynthesis).
- Mesophilic archaea
-
Archaea that grow at temperatures ranging from 20–45 °C.
- Type IV secretion system
-
(T4SS). A secretion system found in many bacteria and some archaea that is involved in the secretion of proteins, DNA and protein–DNA complexes into target cells.
- Hyperthermophile
-
An organism that can thrive at temperatures around 80 °C or higher.
- Nanopods
-
Surface structures that project membrane vesicles from the cell. Nanopods have been found in bacteria and members of the Euryarchaeota.
- Proviruses
-
Viral DNAs that are integrated into the genome of its host.
- Episomes
-
Fragments of DNA that exist independently of the chromosome. Examples of episomes include insertion elements, transposons, plasmids and viruses.
- Temperate viruses
-
Viruses that can integrate into the chromosome or lyse the cell following infection or induction.
- Gene transfer agents
-
A bacteriophage-like element in bacteria that mediates horizontal gene transfer by transducing random host DNA into a recipient cell.
- CRISPR–Cas
-
An adaptive immune system in bacteria and archaea that enables the acquisition of resistance to foreign genetic elements, such as plasmids and viruses.
- Partitioning system
-
A system that ensures the correct segregation of chromosomal DNA or plasmids into the daughter cells of a dividing bacterial cell.
- Hetero-diploid
-
Cells that have two different homologous chromosomes.
- Type IV pilus
-
A surface appendage found in many bacteria and archaea. Type IV pili are involved in motility and attachment to surfaces and other cells.
- Polytopic membrane protein
-
A protein that has multiple transmembrane domains.
Rights and permissions
About this article
Cite this article
Wagner, A., Whitaker, R., Krause, D. et al. Mechanisms of gene flow in archaea. Nat Rev Microbiol 15, 492–501 (2017). https://doi.org/10.1038/nrmicro.2017.41
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2017.41
This article is cited by
-
A self-transmissible plasmid from a hyperthermophile that facilitates genetic modification of diverse Archaea
Nature Microbiology (2023)
-
Hypersaline Lake Urmia: a potential hotspot for microbial genomic variation
Scientific Reports (2023)
-
Bacterial origins of thymidylate metabolism in Asgard archaea and Eukarya
Nature Communications (2023)
-
Symbiotic Interactions of Archaea in Animal and Human Microbiomes
Current Clinical Microbiology Reports (2023)
-
Random transposon mutagenesis identifies genes essential for transformation in Methanococcus maripaludis
Molecular Genetics and Genomics (2023)