Key Points
-
Transcription-coupled repair (TCR) — the fast, preferential repair of the transcribed strand of an active gene — occurs in both prokaryotes and eukaryotes. This kind of repair is performed by the nucleotide excision repair (NER) or the base excision repair (BER) pathways.
-
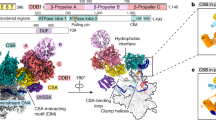
Different factors allow the repair machinery to be specifically targetted to the transcribed strand and the global genome, respectively. The best-studied TCR factors are CSA and CSB (yeast Rad26), but factors such as XPG and TFIIH also have a role.
-
Cells respond to DNA damage by globally downregulating transcription. This might be brought about by several mechanisms working concomitantly.
-
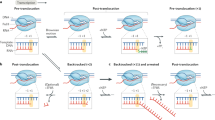
TCR is triggered by the stalled polymerase itself. In the promoter of an active gene, repair is slow. TCR is extra fast immediately downstream from the transcription initiation site, presumably because TFIIH is still associated with the polymerase at this point.
-
Nucleosomes are inhibitory to NER on the non-transcribed strand (repair being slower towards the cores), whereas the speed of TCR is not affected by the presence of the same nucleosomes.
-
In the transcribed strand, RNA polymerase II (RNAPII) is an obstacle to TCR and Rad26/CSB is required to overcome this obstacle to fast repair.
-
Several models for the mechanism of TCR have been proposed, and they all incorporate some sort of displacement of RNAPII from the site of DNA damage.
-
Although much has been learned about the factors and processes affecting TCR, much still needs to be learned about the precise molecular mechanism of the reaction.
Abstract
Several types of helix-distorting DNA lesions block the passage of elongating RNA polymerase II. Surprisingly, such transcription-blocking lesions are usually repaired considerably faster than non-obstructive lesions in the non-transcribed strand or in the genome overall. In this review, our knowledge of eukaryotic transcription-coupled repair (TCR) will be considered from the point of view of transcription, and current models for the mechanism of TCR will be discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tornaletti, S. & Hanawalt, P. C. Effect of DNA lesions on transcription elongation. Biochimie 81, 139–146 (1999).
Mello, J. A., Lippard, S. J. & Essigmann, J. M. DNA adducts of cis-diamminedichloroplatinum(II) and its trans isomer inhibit RNA polymerase II differentially in vivo. Biochemistry 34, 14783–14791 (1995).
Corda, Y., Job, C., Anin, M. F., Leng, M. & Job, D. Spectrum of DNA–platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry 32, 8582–8588 (1993).
Corda, Y., Job, C., Anin, M. F., Leng, M. & Job, D. Transcription by eucaryotic and procaryotic RNA polymerases of DNA modified at a d(GG) or a d(AG) site by the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry 30, 222–230 (1991).
Donahue, B. A., Yin, S., Taylor, J. S., Reines, D. & Hanawalt, P. C. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA 91, 8502–8506 (1994).
Selby, C. P., Drapkin, R., Reinberg, D. & Sancar, A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25, 787–793 (1997).
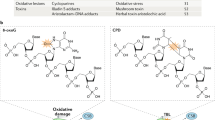
Brooks, P. J. et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 275, 22355–22362 (2000).
Le Page, F. et al. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101, 159–171 (2000).This paper describes the discovery that Cockayne's syndrome mutations lead to a highly elevated mutation frequency at 8-oxo-guanine lesions, and that unrepaired 8-oxo–guanine blocks transcription by RNAPII.
Bohr, V. A., Smith, C. A., Okumoto, D. S. & Hanawalt, P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40, 359–369 (1985).
Mellon, I., Bohr, V. A., Smith, C. A. & Hanawalt, P. C. Preferential DNA repair of an active gene in human cells. Proc. Natl Acad. Sci. USA 83, 8878–8882 (1986).
Mellon, I., Spivak, G. & Hanawalt, P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987).This paper showed that only the transcribed strand of an active gene is preferentially repaired and is therefore the basis for the definition of TCR.
Hanawalt, P. C. Controlling the efficiency of excision repair. Mutat. Res. 485, 3–13 (2001).
Christians, F. C. & Hanawalt, P. C. Repair in ribosomal RNA genes is deficient in xeroderma pigmentosum group C and in Cockayne's syndrome cells. Mutat. Res. 323, 179–187 (1994).
Verhage, R. A., Van de Putte, P. & Brouwer, J. Repair of rDNA in Saccharomyces cerevisiae: RAD4-independent strand-specific nucleotide excision repair of RNA polymerase I transcribed genes Nucleic Acids Res. 24, 1020–1025 (1996).
Balajee, A. S. & Bohr, V. A. Genomic heterogeneity of nucleotide excision repair. Gene 250, 15–30 (2000).
Wood, R. D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 (1997).
Lindahl, T., Karran, P. & Wood, R. D. DNA excision repair pathways. Curr. Opin. Genet. Dev. 7, 158–169 (1997).
Memisoglu, A. & Samson, L. Base excision repair in yeast and mammals. Mutat. Res. 451, 39–51 (2000).
Nouspikel, T., Lalle, P., Leadon, S. A., Cooper, P. K. & Clarkson, S. G. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc. Natl Acad. Sci. USA 94, 3116–3121 (1997).
Cooper, P. K., Nouspikel, T., Clarkson, S. G. & Leadon, S. A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science 275, 990–993 (1997).
Araujo, S. J., Nigg, E. A. & Wood, R. D. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 21, 2281–2291 (2001).
Volker, M. et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224 (2001).
Guzder, S. N., Sung, P., Prakash, L. & Prakash, S. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 271, 8903–8910 (1996).
Svejstrup, J. Q. et al. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 80, 21–28 (1995).
Rodriguez, K. et al. Affinity purification and partial characterization of a yeast multiprotein complex for nucleotide excision repair using histidine-tagged Rad14 protein. J. Biol. Chem. 273, 34180–34189 (1998).
Batty, D., Rapic-Otrin, V., Levine, A. S. & Wood, R. D. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 300, 275–290 (2000).
Sugasawa, K. et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232 (1998).
Svejstrup, J. Q., Vichi, P. & Egly, J. M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem. Sci. 21, 346–350 (1996).
Frit, P., Bergmann, E. & Egly, J. M. Transcription factor IIH: a key player in the cellular response to DNA damage. Biochimie 81, 27–38 (1999).
Henning, K. A. et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82, 555–564 (1995).This paper describes the cloning of the CSA gene.
Troelstra, C. et al. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71, 939–953 (1992).This paper describes the cloning of the CSB gene.
van Hoffen, A. et al. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 21, 5890–5895 (1993).
Venema, J., Mullenders, L. H., Natarajan, A. T., van Zeeland, A. A. & Mayne, L. V. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA 87, 4707–4711 (1990).
Andrews, A. D., Barrett, S. F., Yoder, F. W. & Robbins, J. H. Cockayne's syndrome fibroblasts have increased sensitivity to ultraviolet light but normal rates of unscheduled DNA synthesis. J. Invest. Dermatol. 70, 237–239 (1978).
van Gool, A. J. et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 16, 5955–5965 (1997).
van Gool, A. J. et al. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13, 5361–5369 (1994).
Bhatia, P. K., Verhage, R. A., Brouwer, J. & Friedberg, E. C. Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J. Bacteriol. 178, 5977–5988 (1996).
Eisen, J. A., Sweder, K. S. & Hanawalt, P. C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 (1995).
Citterio, E. et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20, 7643–7653 (2000).
Selby, C. P. & Sancar, A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl Acad. Sci. USA 94, 11205–11209 (1997).This paper describes the finding that CSB affects elongation by RNAPII.
Nakatsu, Y. et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J. Biol. Chem. 275, 34931–34937 (2000).
Drapkin, R. et al. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368, 769–772 (1994).
Schaeffer, L. et al. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science 260, 58–63 (1993).
Schaeffer, L. et al. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 13, 2388–2392 (1994).
Feaver, W. J. et al. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 75, 1379–1387 (1993).
Wang, Z. et al. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature 368, 74–76 (1994).
O'Donovan, A., Davies, A. A., Moggs, J. G., West, S. C. & Wood, R. D. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371, 432–435 (1994).
Habraken, Y., Sung, P., Prakash, L. & Prakash, S. A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J. Biol. Chem. 269, 31342–31345 (1994).
Klungland, A. et al. Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell 3, 33–42 (1999).
Tang, J. Y., Hwang, B. J., Ford, J. M., Hanawalt, P. C. & Chu, G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5, 737–744 (2000).
Venema, J., van Hoffen, A., Natarajan, A. T., van Zeeland, A. A. & Mullenders, L. H. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic Acids Res. 18, 443–448 (1990).
Verhage, R. et al. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 6135–6142 (1994).
Verhage, R. A. et al. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16, 496–502 (1996).This paper shows that yeast cells lacking a TCR gene ( RAD26 ) and a GGR gene ( RAD16 or RAD7 ) have much more defective TCR than cells lacking only RAD26 , which indicates considerable overlap between these repair reactions in yeast.
Critchlow, S. E. & Jackson, S. P. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23, 394–398 (1998).
Yu, A., Fan, H. Y., Liao, D., Bailey, A. D. & Weiner, A. M. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol. Cell 5, 801–810 (2000).
Dianov, G. L., Houle, J. F., Iyer, N., Bohr, V. A. & Friedberg, E. C. Reduced RNA polymerase II transcription in extracts of cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 25, 3636–3642 (1997).
Balajee, A. S., May, A., Dianov, G. L., Friedberg, E. C. & Bohr, V. A. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA 94, 4306–4311 (1997).
van Oosterwijk, M. F., Versteeg, A., Filon, R., van Zeeland, A. A. & Mullenders, L. H. The sensitivity of Cockayne's syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol. Cell. Biol. 16, 4436–4444 (1996).
Rockx, D. A. et al. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl Acad. Sci. USA 97, 10503–10508 (2000).
Dahmus, M. E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271, 19009–19012 (1996).
Vichi, P. et al. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 16, 7444–7456 (1997).
You, Z., Feaver, W. J. & Friedberg, E. C. Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26. Mol. Cell. Biol. 18, 2668–2676 (1998).
Christians, F. C. & Hanawalt, P. C. Inhibition of transcription and strand-specific DNA repair by α-amanitin in Chinese hamster ovary cells. Mutat. Res. 274, 93–101 (1992).
Sweder, K. S. & Hanawalt, P. C. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl Acad. Sci. USA 89, 10696–10700 (1992).
Teng, Y. & Waters, R. Excision repair at the level of the nucleotide in the upstream control region, the coding sequence and in the region where transcription terminates of the Saccharomyces cerevisiae MFA2 gene and the role of RAD26. Nucleic Acids Res. 28, 1114–1119 (2000).
Tijsterman, M. & Brouwer, J. Rad26, the yeast homolog of the cockayne syndrome B gene product, counteracts inhibition of DNA repair due to RNA polymerase II transcription. J. Biol. Chem. 274, 1199–1202 (1999).This paper indicates that a major role of Rad26/CSB is to allow repair of DNA damage encountered by elongating RNAPII.
Tijsterman, M., de Pril, R., Tasseron-de Jong, J. G. & Brouwer, J. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol. 19, 934–940 (1999).
Tijsterman, M., Verhage, R. A., van de Putte, P., Tasseron-de Jong, J. G. & Brouwer, J. Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 94, 8027–8032 (1997).
Tijsterman, M., Tasseron-de Jong, J. G., van de Putte, P. & Brouwer, J. Transcription-coupled and global genome repair in the Saccharomyces cerevisiae RPB2 gene at nucleotide resolution. Nucleic Acids Res. 24, 3499–3506 (1996).
Tu, Y., Tornaletti, S. & Pfeifer, G. P. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J. 15, 675–683 (1996).
Tu, Y., Bates, S. & Pfeifer, G. P. Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J. Biol. Chem. 272, 20747–20755 (1997).
Tu, Y., Bates, S. & Pfeifer, G. P. The transcription-repair coupling factor CSA is required for efficient repair only during the elongation stages of RNA polymerase II transcription. Mutat. Res. 400, 143–151 (1998).
Dvir, A., Conaway, J. W. & Conaway, R. C. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. 11, 209–214 (2001).
Zawel, L., Kumar, K. P. & Reinberg, D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9, 1479–1490 (1995).
Tantin, D., Kansal, A. & Carey, M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 17, 6803–6814 (1997).
Tantin, D. RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J. Biol. Chem. 273, 27794–27799 (1998).
Wellinger, R. E. & Thoma, F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 16, 5046–5056 (1997).
Wada, T. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998).
Hartzog, G. A., Wada, T., Handa, H. & Winston, F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12, 357–369 (1998).
Jansen, L. E. et al. Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J. 19, 6498–6507 (2000).
Otero, G. et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118 (1999).
Svejstrup, J. Q. Transcription-coupled DNA repair without the transcription–coupling repair factor. Trends Biochem. Sci. 26, 151 (2001).
Selby, C. P. & Sancar, A. Molecular mechanism of transcription-repair coupling. Science 260, 53–58 (1993).
Selby, C. P. & Sancar, A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 272, 1885–1890 (1997).
Hara, R., Selby, C. P., Liu, M., Price, D. H. & Sancar, A. Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 274, 24779–24786 (1999).
Verhage, R. A., Heyn, J., van de Putte, P. & Brouwer, J. Transcription elongation factor S-II is not required for transcription-coupled repair in yeast. Mol. Gen. Genet. 254, 284–290 (1997).
Peterson, C. L. & Workman, J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10, 187–192 (2000).
Mu, D. & Sancar, A. Model for XPC-independent transcription-coupled repair of pyrimidine dimers in humans. J. Biol. Chem. 272, 7570–7573 (1997).
Bregman, D. B. et al. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA 93, 11586–11590 (1996).This paper shows that RNAPII is post-translationally modified in response to UV-irradiation, and that this modification is compromised in Cockayne's syndrome cells.
Beaudenon, S. L., Huacani, M. R., Wang, G., McDonnell, D. P. & Huibregtse, J. M. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6972–6979 (1999).
Huibregtse, J. M., Yang, J. C. & Beaudenon, S. L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl Acad. Sci. USA 94, 3656–3661 (1997).
Ratner, J. N., Balasubramanian, B., Corden, J., Warren, S. L. & Bregman, D. B. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 273, 5184–5189 (1998).
McKay, B. C. et al. UV light-induced degradation of RNA polymerase II is dependent on the Cockayne's syndrome A and B proteins but not p53 or MLH1. Mutat. Res. 485, 93–105 (2001).
Luo, Z., Zheng, J., Lu, Y. & Bregman, D. B. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 486, 259–274 (2001).
Lommel, L., Bucheli, M. E. & Sweder, K. S. Transcription-coupled repair in yeast is independent from ubiquitylation of RNA pol II: implications for Cockayne's syndrome. Proc. Natl Acad. Sci. USA 97, 9088–9092 (2000).
Friedberg, E. C., Fischhaber, P. L. & Kisker, C. Error-prone DNA polymerases. Novel structures and the benefits of infidelity. Cell 107, 9–12 (2001).
Holbrook, N. J. & Fornace, A. J. Jr. Response to adversity: molecular control of gene activation following genotoxic stress. New Biol. 3, 825–833 (1991).
de Boer, J. & Hoeijmakers, J. H. Nucleotide excision repair and human syndromes. Carcinogenesis 21, 453–460 (2000).Excellent review of syndromes that are associated with defects in DNA repair and TCR.
Mayne, L. V. & Lehmann, A. R. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 42, 1473–1478 (1982).
Lehmann, A. R. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15, 15–23 (2001).
Berneburg, M. & Lehmann, A. R. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet 43, 71–102 (2001).
Hanawalt, P. C. DNA repair. The bases for Cockayne syndrome. Nature 405, 415–416 (2000).
Citterio, E., Vermeulen, W. & Hoeijmakers, J. H. Transcriptional healing. Cell 101, 447–450 (2000).
Acknowledgements
Work relevant to this review was supported by an in-house grant from the Imperial Cancer Research Fund. T. Lindahl and B. Winkler are thanked for their comments and suggestions.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Encyclopedia of Life Sciences:
DNA-repair-deficiency disorders
Saccharomyces Genome Database:
FURTHER READING
Glossary
- DNA LIGASE
-
Seals DNA breaks containing a 5′-phosphorylated end and a 3′-OH end. Several different DNA ligases are found in eukaryotic cells.
- TFIIH
-
(Transcription factor IIH). One of the factors that are generally required for RNA polymerase II-dependent transcription. Also has an essential role in nucleotide excision repair.
- HELICASE
-
An enzyme that uses the energy of ATP hydrolysis to unwind/separate the two strands of DNA.
- ENDONUCLEASE
-
Cuts DNA internally. Often, but not always, cuts at specific recognition sequences.
- DNA GLYCOSYLASE
-
Binds specifically to a target base and hydrolyses the N-glycosylic bond. This releases the inappropriate base while keeping the DNA backbone intact. Several distinct glycosylases exist in eukaryotic cells.
- ABASIC SITE
-
A nucleotide site that has lost the base.
- 'FLAP' ENDONUCLEASE
-
A structure-specific endonuclease, which cleaves off a single-stranded piece of DNA that is not base-paired owing to, for example, mismatched bases.
- SNF2 PROTEIN FAMILY
-
A family of proteins that have helicase-like ATPase domains, yet are not helicases. Rather than unwinding DNA strands, these proteins remodel protein–DNA contacts, such as histone–DNA contacts.
- SWI/SNF-LIKE COMPLEXES
-
Protein complexes that contain a subunit that is homologous to SNF2.
- WD40 REPEAT
-
A protein motif used by a variety of proteins for protein–protein interactions.
- ORTHOLOGUE
-
A homologue whose sequence and function has been conserved during evolution.
- RING FINGER
-
Protein motif that is found in ubiquitin ligases.
- RAD7 AND RAD16
-
Proteins that are required for global genome repair in yeast. They are very important for nucleotide excision repair of the non-transcribed strand of an active gene, but have only a minor role for repair on the transcribed strand/transcription-coupled repair.
- NON-HOMOLOGOUS END-JOINING
-
Repair pathway in which severed DNA ends are re-joined. The prominent pathway for repairing double-stranded breaks in higher eukaryotic cells.
- HYPOPHOSPHORYLATED RNAPII A-FORM
-
The carboxy-terminal domain (CTD) of the largest RNAPII subunit comprises 26–52 heptapeptide repeats (consensus Tyr-Ser-Pro-Thr-Ser-Pro-Ser). The RNAPII form that is hypophosphorylated on the CTD is called RNAPII-A, whereas the form that is multiply phosphorylated (hyperphosphorylated) on Ser2 and Ser5 of the repeat is called RNAPII-0.
- TATA-BINDING PROTEIN
-
The TATA-binding protein (TBP) binds directly to the TATA sequence in the core promoter of a gene to initiate assembly of the general transcription factor machinery that enables RNAPII to transcribe the gene.
- UBIQUITYLATION
-
The covalent linkage of the small protein ubiquitin to other proteins is essential for a variety of cellular processes. Ubiquitylation can mark proteins for rapid intracellular degradation, but can also have other consequences for the modified substrate protein.
- UBIQUITIN LIGASE
-
The last enzyme in a cascade of ubiquitin-mobilizing proteins that together enable ubiquitylation. First, ubiquitin is activated by formation of a thioester bond to a ubiquitin- activating enzyme (E1). After transfer to a ubiquitin- conjugating enzyme (E2), ubiquitin is ligated to a protein substrate with the help of a ubiquitin protein ligase (E3). Several ubiquitin moieties can be added to the substrate, each time forming an isopeptide bond to an internal lysine residue.
Rights and permissions
About this article
Cite this article
Svejstrup, J. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol 3, 21–29 (2002). https://doi.org/10.1038/nrm703
Issue Date:
DOI: https://doi.org/10.1038/nrm703
This article is cited by
-
Associations between nucleosome phasing, sequence asymmetry, and tissue-specific expression in a set of inbred Medaka species
BMC Genomics (2015)
-
An in vitro method for detecting genetic toxicity based on inhibition of RNA synthesis by DNA lesions
Genes and Environment (2015)
-
Association of purine asymmetry, strand-biased gene distribution and PolC within Firmicutes and beyond: a new appraisal
BMC Genomics (2014)
-
Substitution rate variation at human CpG sites correlates with non-CpG divergence, methylation level and GC content
Genome Biology (2011)
-
The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2to maintain genome integrity upon UV stress
The EMBO Journal (2011)