Key Points
-
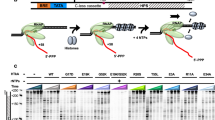
Histone exchange involves the partial or complete exchange of nucleosomes for newer or altered components. This process occurs sequentially through the removal and the replacement of the H2A–H2B dimers followed by the H3–H4 tetramer.
-
Several factors that affect the stability of the nucleosome influence the process of histone exchange. These include chromatin modifiers, chromatin remodellers and histone chaperones.
-
Destabilization of the nucleosome allows histone exchange to proceed, often resulting in the replacement of canonical histones with variants that carry out specialized cellular functions.
-
Histone exchange features prominently during the process of transcription initiation and elongation. A combination of variant exchange and turnover of histone subunits drives RNA polymerase II (Pol II)-mediated transcription.
-
Resetting of chromatin is a crucial process used by the cell to reassemble the nucleosomes that are lost during the transcription process. The co-transcriptional histone H3 lysine 36 methylation mark uses a multipronged approach to prevent histone exchange over coding regions.
-
Limiting unobstructed histone exchange over coding regions of genes is necessary to prevent aberrant initiation of transcription. Given the importance of non-coding RNA in the development of diseases, understanding how they are produced has immense value.
Abstract
The packaging of DNA into strings of nucleosomes is one of the features that allows eukaryotic cells to tightly regulate gene expression. The ordered disassembly of nucleosomes permits RNA polymerase II (Pol II) to access the DNA, whereas nucleosomal reassembly impedes access, thus preventing transcription and mRNA synthesis. Chromatin modifications, chromatin remodellers, histone chaperones and histone variants regulate nucleosomal dynamics during transcription. Disregulation of nucleosome dynamics results in aberrant transcription initiation, producing non-coding RNAs. Ongoing research is elucidating the molecular mechanisms that regulate chromatin structure during transcription by preventing histone exchange, thereby limiting non-coding RNA expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jonkers, I. & Lis, J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nature Rev. Mol. Cell Biol. http://dx.doi.org/10.1038/nrm3953 (2015).
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nature Rev. Mol. Cell Biol. http://dx.doi.org/10.1038/nrm3952 (2015).
Pourra, O. & Domenico Libri, D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nature Rev. Mol. Cell Biol. http://dx.doi.org/10.1038/nrm3943 (2015).
Luger, K., Dechassa, M. L. & Tremethick, D. J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nature Rev. Mol. Cell Biol. 13, 436–447 (2012).
Talbert, P. B. & Henikoff, S. Histone variants — ancient wrap artists of the epigenome. Nature Rev. Mol. Cell Biol. 11, 264–275 (2010).
Kornberg, R. D. Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 (1974).
Marzluff, W. F., Gongidi, P., Woods, K. R., Jin, J. & Maltais, L. J. The human and mouse replication-dependent histone genes. Genomics 80, 487–498 (2002).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Weber, C. M. & Henikoff, S. Histone variants: dynamic punctuation in transcription. Genes Dev. 28, 672–682 (2014).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). Purification of the human variant histones carried out in this study helped to define the novel interaction profiles, particularly with respect to histone chaperones.
Stoler, S., Keith, K. C., Curnick, K. E. & Fitzgerald-Hayes, M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573–586 (1995).
Schenk, R., Jenke, A., Zilbauer, M., Wirth, S. & Postberg, J. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 (2011).
Montellier, E. et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 27, 1680–1692 (2013).
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). This is the first report of the nucleosome crystal structure, which underscores the importance of a modular organization.
Smith, S. & Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991).
Kulaeva, O. I., Hsieh, F. K. & Studitsky, V. M. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl Acad. Sci. USA 107, 11325–11330 (2010).
Jamai, A., Imoberdorf, R. M. & Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007).
Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Rev. Genet. 9, 15–26 (2008).
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E. & Tyler, J. K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Tachiwana, H. et al. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. D Biol. Crystallogr. 67, 578–583 (2011).
Elsasser, S. J. et al. DAXX envelops a histone H3.3–H4 dimer for H3.3specific recognition. Nature 491, 560–565 (2012).
Liu, C. P. et al. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nature Struct. Mol. Biol. 19, 1287–1292 (2012).
Mito, Y., Henikoff, J. G. & Henikoff, S. Genome-scale profiling of histone H3.3 replacement patterns. Nature Genet. 37, 1090–1097 (2005).
Wirbelauer, C., Bell, O. & Schubeler, D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19, 1761–1766 (2005).
Nakayama, T., Nishioka, K., Dong, Y. X., Shimojima, T. & Hirose, S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 21, 552–561 (2007).
Daury, L., Chailleux, C., Bonvallet, J. & Trouche, D. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 7, 66–71 (2006).
Mito, Y., Henikoff, J. G. & Henikoff, S. Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411 (2007).
Jin, C. et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nature Genet. 41, 941–945 (2009). This study defines the genomic localization of the fragile double-variant nucleosomes.
Kraushaar, D. C. et al. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 (2013).
Huang, C. et al. H3.3–H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 9, e1003558 (2013).
Deal, R. B., Henikoff, J. G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010). This paper from defines a novel method that uses metabolic labelling to follow histone turnover in metazoans.
Chow, C. M. et al. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 6, 354–360 (2005).
Filipescu, D., Szenker, E. & Almouzni, G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 29, 630–640 (2013).
Szenker, E., Ray-Gallet, D. & Almouzni, G. The double face of the histone variant H3.3. Cell Res. 21, 421–434 (2011).
Elsaesser, S. J., Goldberg, A. D. & Allis, C. D. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20, 110–117 (2010).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A. Z. Nature Struct. Biol. 7, 1121–1124 (2000).
Meneghini, M. D., Wu, M. & Madhani, H. D. Conserved histone variant H2A. Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 (2003).
Obri, A. et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014).
Smolle, M. & Workman, J. L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta 1829, 84–97 (2013).
Petruk, S. et al. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150, 922–933 (2012).
Williams, S. K., Truong, D. & Tyler, J. K. Acetylation in the globular core of histone H3 on lysine 56 promotes chromatin disassembly during transcriptional activation. Proc. Natl Acad. Sci. USA 105, 9000–9005 (2008).
Tropberger, P. et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152, 859–872 (2013).
Shogren-Knaak, M. et al. Histone H4K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Lee, J. S. et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131, 1084–1096 (2007).
Venkatesh, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J. & Peterson, C. L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 (2011). This paper characterizes the role of the INO80 complex in preventing the mislocalization of the variant H2A.Z in genomic regions other than the promoter.
McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004).
Tropberger, P. & Schneider, R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nature Struct. Mol. Biol. 20, 657–661 (2013).
Tessarz, P. & Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nature Rev. Mol. Cell Biol. 15, 703–708 (2014).
Becker, P. B. & Workman, J. L. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5, a017905 (2013).
Kobor, M. S. et al. A protein complex containing the conserved Swi2/Snf2related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131 (2004).
Watanabe, S. & Peterson, C. L. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb. Symp. Quant. Biol. 75, 35–42 (2010).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Luk, E. et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 (2010).
Wu, W. H. et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nature Struct. Mol. Biol. 12, 1064–1071 (2005).
Watanabe, S., Radman-Livaja, M., Rando, O. J. & Peterson, C. L. A histone acetylation switch regulates H2A.Z deposition by the SWRC remodeling enzyme. Science 340, 195–199 (2013).
Kusch, T. et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 (2004).
Ruhl, D. D. et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45, 5671–5677 (2006).
Eissenberg, J. C., Wong, M. & Chrivia, J. C. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the notch signaling pathway. Mol. Cell. Biol. 25, 6559–6569 (2005).
Cai, Y. et al. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280, 13665–13670 (2005).
Johnston, H., Kneer, J., Chackalaparampil, I., Yaciuk, P. & Chrivia, J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 274, 16370–16376 (1999).
Conaway, R. C. & Conaway, J. W. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 34, 71–77 (2009).
Yen, K., Vinayachandran, V. & Pugh, B. F. SWRC and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256 (2013).
D'Arcy, S. et al. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A–H2B in an unconventional tetrameric form. Mol. Cell 51, 662–677 (2013).
De Koning, L., Corpet, A., Haber, J. E. & Almouzni, G. Histone chaperones: an escort network regulating histone traffic. Nature Struct. Mol. Biol. 14, 997–1007 (2007).
Hondele, M. et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013).
Bowman, A. et al. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol. Cell 41, 398–408 (2011).
Swaminathan, V., Kishore, A. H., Febitha, K. K. & Kundu, T. K. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 25, 7534–7545 (2005).
Luk, E. et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25, 357–368 (2007).
Su, D. et al. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature 483, 104–107 (2012).
Owen-Hughes, T. & Workman, J. L. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 15, 4702–4712 (1996).
Lorch, Y., Maier-Davis, B. & Kornberg, R. D. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl Acad. Sci. USA 103, 3090–3093 (2006).
Kuryan, B. G. et al. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc. Natl Acad. Sci. USA 109, 1931–1936 (2012).
Han, J. et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315, 653–655 (2007).
Driscoll, R., Hudson, A. & Jackson, S. P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 (2007).
Rufiange, A., Jacques, P. E., Bhat, W., Robert, F. & Nourani, A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007).
Schwabish, M. A. & Struhl, K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22, 415–422 (2006).
Kolonko, E. M. et al. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc. Natl Acad. Sci. USA 107, 20275–20280 (2010).
Tsubota, T. et al. Histone H3K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25, 703–712 (2007).
Kaplan, T. et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4, e1000270 (2008).
Youdell, M. L. et al. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28, 4915–4926 (2008).
Yoh, S. M., Cho, H., Pickle, L., Evans, R. M. & Jones, K. A. The Spt6 SH2 domain binds Ser2P RNAPII to direct Iws1dependent mRNA splicing and export. Genes Dev. 21, 160–174 (2007).
Du, H. N., Fingerman, I. M. & Briggs, S. D. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes Dev. 22, 2786–2798 (2008).
Du, H. N. & Briggs, S. D. A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys36 methylation, histone acetylation, and repression of cryptic transcription. J. Biol. Chem. 285, 11704–11713 (2010).
Carrozza, M. J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Kaplan, C. D., Laprade, L. & Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 (2003).
Hassan, A. H., Neely, K. E. & Workman, J. L. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104, 817–827 (2001).
Schneiderman, J. I., Orsi, G. A., Hughes, K. T., Loppin, B. & Ahmad, K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl Acad. Sci. USA 109, 19721–19726 (2012).
Drew, H. R. & Travers, A. A. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 186, 773–790 (1985).
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006).
Hartley, P. D. & Madhani, H. D. Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458 (2009).
Ganguli, D., Chereji, R. V., Iben, J. R., Cole, H. A. & Clark, D. J. RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Res. 24, 1637–1649 (2014).
Ranjan, A. et al. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232–1245 (2013). References 54–56 and 94 define the factors regulating the function of SWR in replacing histone variant H2A.Z.
Yuan, G. C. et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 (2005).
Mavrich, T. N. et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18, 1073–1083 (2008).
Zofall, M. et al. Histone H2A.Z cooperates with RNAi heterochromatin factors suppress antisense RNAs. Nature 461, 419–422 (2009).
Zhang, H., Roberts, D. N. & Cairns, B. R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231 (2005).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007). References 77 and 99 define the concept, genomic distribution and factors governing histone turnover in wild-type yeast.
Durant, M. & Pugh, B. F. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 27, 5327–5335 (2007).
Altaf, M. et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A. Z by the SWR1 complex. J. Biol. Chem. 285, 15966–15977 (2010).
Halley, J. E., Kaplan, T., Wang, A. Y., Kobor, M. S. & Rine, J. Roles for H2A. Z and its acetylation in GAL1 transcription and gene induction, but not GAL1transcriptional memory. PLoS Biol. 8, e1000401 (2010).
Draker, R. et al. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 8, e1003047 (2012).
Chen, P. et al. H3.3 actively marks enhancers and primes gene transcription via opening higher- ordered chromatin. Genes Dev. 27, 2109–2124 (2013).
Heinz, S. et al. The selection and function of cell type-specific enhancers. Nature Rev. Mol. Cell Biol. http://dx.doi.org/10.1038/nrm3949 (2015).
Conerly, M. L. et al. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 20, 1383–1390 (2010).
Zilberman, D., Coleman-Derr, D., Ballinger, T. & Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008).
Pchelintsev, N. A. et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019 (2013).
Kireeva, M. L. et al. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002).
Kulaeva, O. I. et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nature Struct. Mol. Biol. 16, 1272–1278 (2009).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Pavri, R. et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 (2006).
Hsieh, F. K. et al. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl Acad. Sci. USA 110, 7654–7659 (2013).
Sarai, N. et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 32, 2392–2406 (2013).
Kulaeva, O. I., Hsieh, F. K., Chang, H. W., Luse, D. S. & Studitsky, V. M. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta 1829, 76–83 (2013).
Fleming, A. B., Kao, C. F., Hillyer, C., Pikaart, M. & Osley, M. A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 (2008).
Joshi, A. A. & Struhl, K. Eaf3 chromodomain interaction with methylated H3K36 links histone deacetylation to Pol II elongation. Mol. Cell 20, 971–978 (2005).
Cheung, V. et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6, e277 (2008). This study emphasizes the role of chromatin in regulating transcription from cryptic promoters.
Gossett, A. J. & Lieb, J. D. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet. 8, e1002771 (2012).
Yoh, S. M., Lucas, J. S. & Jones, K. A. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2mediated histone H3K36 methylation. Genes Dev. 22, 3422–3434 (2008).
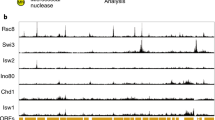
Smolle, M. et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nature Struct. Mol. Biol. 19, 884–892 (2012). References 46 and 121 define the mechanism involved in resetting chromatin after transcription.
Winkler, D. D., Muthurajan, U. M., Hieb, A. R. & Luger, K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 (2011).
Carvalho, S. et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, 2881–2893 (2013).
Maltby, V. E. et al. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 32, 3479–3485 (2012).
Radman-Livaja, M. et al. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS Genet. 8, e1002811 (2012).
Hennig, B. P., Bendrin, K., Zhou, Y. & Fischer, T. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 13, 997–1003 (2012).
Lee, J. S. et al. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 26, 914–919 (2012).
Batta, K., Zhang, Z., Yen, K., Goffman, D. B. & Pugh, B. F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 (2011).
Govind, C. K. et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39, 234–246 (2010).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).
Ooi, S. L., Priess, J. R. & Henikoff, S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2, e97 (2006).
Loyola, A. & Almouzni, G. Marking histone H3 variants: how, when and why? Trends Biochem. Sci. 32, 425–433 (2007).
Wen, H. et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014).
Guo, R. et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 (2014). References 133 and 134 define the identification of a human protein, ZMYND11, that specifically recognizes the H3K36-methylated histone variant H3.3 and that is involved in Pol II elongation and RNA splicing.
Wen, H., Li, Y., Li, H. & Shi, X. ZMYND11: an H3.3-specific reader of H3K36me3. Cell Cycle 13, 2153–2154 (2014).
Jelinic, P., Pellegrino, J. & David, G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol. Cell. Biol. 31, 54–62 (2011).
Kumar, G. S. et al. Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J. Mol. Biol. 422, 519–531 (2012).
Li, B. et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 (2007).
Esteller, M. Non-coding RNAs in human disease. Nature Rev. Genet. 12, 861–874 (2011).
Zovoilis, A. et al. The expression level of small non-coding RNAs derived from the first exon of protein-coding genes is predictive of cancer status. EMBO Rep. 15, 402–410 (2014).
Jiang, L. et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227 (2013).
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Pradeepa, M. M., Sutherland, H. G., Ule, J., Grimes, G. R. & Bickmore, W. A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717 (2012). References 142 and143 enumerate the role of chromatin structure and modification in co-transcriptional regulation of alternative splicing.
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R. & Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 41, 376–381 (2009).
Bartolomei, M. S., Halden, N. F., Cullen, C. R. & Corden, J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol. 8, 330–339 (1988).
Litingtung, Y. et al. Growth retardation and neonatal lethality in mice with a homozygous deletion in the Cterminal domain of RNA polymerase II. Mol. Gen. Genet. 261, 100–105 (1999).
Hsin, J. P. & Manley, J. L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012).
Heidemann, M., Hintermair, C., Voss, K. & Eick, D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 1829, 55–62 (2013).
Smolle, M. & Venkatesh, S. in Fundamentals of Chromatin (eds Workman, J. L. & Abmayr, S. M.) (Springer, 2014).
Deal, R. B. & Henikoff, S. Capturing the dynamic epigenome. Genome Biol. 11, 218 (2010).
Kimura, H. & Cook, P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341–1353 (2001).
Voss, T. C. & Hager, G. L. Visualizing chromatin dynamics in intact cells. Biochim. Biophys. Acta 1783, 2044–2051 (2008).
Misteli, T., Gunjan, A., Hock, R., Bustin, M. & Brown, D. T. Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 (2000).
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).
Verzijlbergen, K. F. et al. Recombination-induced tag exchange to track old and new proteins. Proc. Natl Acad. Sci. USA 107, 64–68 (2010).
Xu, M. et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 (2010).
Sweet, S. M., Li, M., Thomas, P. M., Durbin, K. R. & Kelleher, N. L. Kinetics of reestablishing H3K79 methylation marks in global human chromatin. J. Biol. Chem. 285, 32778–32786 (2010).
Venkatesh, S. & Workman, J. L. Recognizing methylated histone variant H3.3 to prevent tumors. Cell Res. 24, 649–650 (2014).
Kang, B. et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 25, 1359–1364 (2011).
Straube, K., Blackwell, J. S. Jr & Pemberton, L. F. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic 11, 185–197 (2010).
Imbeault, D., Gamar, L., Rufiange, A., Paquet, E. & Nourani, A. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 283, 27350–27354 (2008).
Mao, Z. et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A. Z. Cell Res. 24, 389–399 (2014).
Acknowledgements
The authors thank K. Natarajan for critical reading of this manuscript and apologize to several colleagues whose work could not be cited here because of space restrictions. This work was supported in part by the US National Institutes of Health (NIH) grant NIH R01GM047867 to J.L.W. and the Stowers Institute for Medical Research.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Histone variants
-
Histone protein isoforms transcribed from distinct genomic loci that can replace the typical histone proteins in a defined manner in order to achieve specialized functions.
- Linker histone H1
-
A component of the nucleosome that is necessary to lock the DNA wrapped around the histone core at the dyad axis and to contribute to the higher-order structure of chromatin. It does not contain the histone-fold domain and is not a part of the globular core of the nucleosome.
- Chromosomal domain segregation
-
The separation of chromatin, on the basis of the packing density, into euchromatin (less dense) and heterochromatin (more dense) domains, which are separated by distinct chromatin structures that often involve the H2A.Z histone variant.
- Ubiquitylation
-
A post-translational modification on histones that adds a single ubiquitin protein molecule to lysine residues, resulting in transcriptional regulation. This is different from the addition of a chain of ubiquitin molecules, which usually signals for degradation.
- Sumoylation
-
A post-translational modification similar to ubiquitylation, in which a small ubiquitin-like modifier (SUMO) protein is added to lysine residues to regulate transcription. In contrast to ubiquitylation, sumoylation is not involved in protein degradation.
- Open chromatin
-
A qualitative term applied to the fluid state of chromatin that allows easy access to the DNA; it is usually associated with active post-translational modification marks.
- Closed chromatin
-
A qualitative term applied to the static state of chromatin that prevents access to the DNA because of the addition of repressive post-translational modification marks and the binding of factors that pack nucleosomes into a compact structure.
- Readers
-
Dedicated protein factors that have the ability, through specialized domains, to recognize either specific post-translational marks on histones or a combination of marks and histone variants to direct a particular transcriptional outcome.
- Writers
-
Enzymes that add post-translational modifications on histones. Each is specific for a particular class of post-translational modification (for example, kinases for phosphorylation and methyltransferases for methylation).
- Erasers
-
Enzymes that remove specific post-translational modification marks from histone substrates and that belong to various classes (for example, phosphatases for dephosphosphorylation and demethylases for demethylation).
- DNA translocase
-
An enzyme that catalyses the ATP-dependent breakage of histone–DNA contacts and the subsequent pushing of the dislocated DNA segment, which results in its movement around a central anchored nucleosome.
- SNF2 family
-
A large family of DNA-dependent ATPases, which are helicase-like proteins involved in chromatin remodelling. The family was identified on the basis of protein sequence homology to the ATPase domain of yeast Snf2.
- Histone acetyltransferase
-
An enzyme that catalyses the addition of an acetyl moiety from the donor acetyl coenzyme A to lysine residues on histones.
- Histone sinks
-
Proteins (often histone chaperones) that accept histone subunits removed from nucleosomes, which prevent their nonspecific association with DNA.
- Histone methyltransferase
-
An enzyme that catalyses the addition of up to three methyl moieties from the donor S-adenosyl methionine to lysine residues on histone proteins.
- PWWP domain
-
A 135 amino acid protein domain characterized by a central proline-tryptophan-tryptophan-proline core, that recognizes methylated lysines (particularly H3 K36) as a part of several eukaryotic proteins involved in chromatin regulation, DNA repair and transcriptional regulation.
Rights and permissions
About this article
Cite this article
Venkatesh, S., Workman, J. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16, 178–189 (2015). https://doi.org/10.1038/nrm3941
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3941