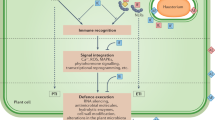
Key Points
-
An overview of the various mechanisms of bacterial pathogenesis is presented, with a focus on bacterial proteins, protein-secretion systems and small molecules that modulate plant biology.
-
The mechanisms by which plant pathogen recognition receptors (PRRs) recognize bacterial pathogen-associated molecular patterns (PAMPS) are discussed. Latest developments in this field are examined with an emphasis on the flagellin–FLS2 and elongation factor Tu (EF-Tu)–EFR (EF-Tu receptor) systems.
-
PRR-mediated defences represent a significant obstacle for plant pathogens. However, type III effectors that function to suppress PRR-mediated basal defences have been discovered. These effectors have been shown to limit cell wall-based defences and expression of defence-associated genes.
-
Plants probably evolved resistance (R) proteins to recognize the presence of bacterial effectors that suppress PRR-mediated defences. R proteins elicit a strong hypersensitive response that provides immunity to the host plant. Mechanisms of effector–R protein recognition are discussed in the context of current models.
-
In response to plant R proteins, it seems that pathogens have evolved an alternative set of effectors that suppress R-protein-mediated defences. The mechanisms by which effectors suppress R-protein-mediated defences are examined.
-
Current research establishes that resistance to bacterial pathogens is regulated by both PRR- and R-protein-mediated recognition. The outcome of each interaction is dependent on at least four factors: the complement of PAMPS and effectors in the pathogen, and the PRR and R proteins in the host.
Abstract
Recent research on plant responses to bacterial attack has identified extracellular and intracellular host receptors that recognize conserved pathogen-associated molecular patterns and more specialized virulence proteins, respectively. These findings have shed light on our understanding of the molecular mechanisms by which bacteria elicit host defences and how pathogens have evolved to evade or suppress these defences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Genin, S. & Boucher, C. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42, 107–134 (2004).
Nino-Liu, D. O., Ronald, P. R. & Bogdanove, A. J. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. (in the press).
Preston, G. M. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol. Plant Pathol. 1, 263–275 (2000).
Van Sluys, M. A. et al. Comparative genomic analysis of plant-associated bacteria. Annu. Rev. Phytopathol. 40, 169–189 (2002).
Agrios, G. N. Plant Pathology (Academic Press, San Diego, 1997).
Anonymous. Florida citrus canker eradication program. University of Florida, Institute of Food and Agricultural Sciences [online] (2006).
Brown, K. Florida fights to stop citrus canker. Science 292, 2275–2276 (2001).
Strange, R. N. & Scott, P. R. Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116 (2005).
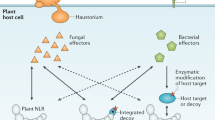
Buttner, D. & Bonas, U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr. Opin. Microbiol. 9, 193–200 (2006).
Mudgett, M. B. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531 (2005).
Alfano, J. R. & Collmer, A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414 (2004).
Chang, J. H. et al. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl Acad. Sci. USA 102, 2549–2554 (2005).
Preston, G. M., Studholme, D. J. & Caldelari, I. Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol. Rev. 29, 331–360 (2005).
Jha, G., Rajeshwari, R. & Sonti, R. V. Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 18, 891–898 (2005).
Toth, I. K. & Birch, P. R. Rotting softly and stealthily. Curr. Opin. Plant Biol. 8, 424–429 (2005).
Nomura, K., Melotto, M. & He, S. Y. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8, 361–368 (2005).
Petnicki-Ocwieja, T. et al. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA 99, 7652–7657 (2002).
Shao, F., Merritt, P. M., Bao, Z., Innes, R. W. & Dixon, J. E. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109, 575–588 (2002).
Coaker, G., Falick, A. & Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308, 548–550 (2005). This study, along with reference 20, shows that the type III effector AvrRpt2 cleaves the plant defence regulator RIN4.
Kim, H. S. et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl Acad. Sci. USA 102, 6496–6501 (2005).
Lopez-Solanilla, E., Bronstein, P. A., Schneider, A. R. & Collmer, A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54, 353–365 (2004).
Hotson, A., Chosed, R., Shu, H., Orth, K. & Mudgett, M. B. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50, 377–389 (2003).
Roden, J., Eardley, L., Hotson, A., Cao, Y. & Mudgett, M. B. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17, 633–643 (2004).
Abramovitch, R. B., Janjusevic, R., Stebbins, C. E. & Martin, G. B. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl Acad. Sci. USA 103, 2851–2856 (2006).
Janjusevic, R., Abramovitch, R. B., Martin, G. B. & Stebbins, C. E. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226 (2006). References 24 and 25 show that AvrPtoB has E3 ubiquitin ligase activity that is required to suppress plant defences.
Bretz, J. R. et al. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 49, 389–400 (2003).
Espinosa, A., Guo, M., Tam, V. C., Fu, Z. Q. & Alfano, J. R. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49, 377–387 (2003).
Nimchuk, Z. et al. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363 (2000).
Shan, L., Thara, V. K., Martin, G. B., Zhou, J. M. & Tang, X. The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12, 2323–2338 (2000).
Robert-Seilaniantz, A., Shan, L., Zhou, J. M. & Tang, X. The Pseudomonas syringae pv. tomato DC3000 Type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19, 130–138 (2006).
Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S. & Cascales, E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 (2005).
Tzfira, T., Vaidya, M. & Citovsky, V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92 (2004).
Gelvin, S. B. Agrobacterium and plant genes involved in t-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223–256 (2000).
da Silva, A. C. et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463 (2002).
Bender, C. L., Alarcon-Chaidez, F. & Gross, D. C. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292 (1999).
Glickmann, E. et al. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 11, 156–162 (1998).
Navarro, L. et al. A plant MiRNA contributes to antibacterial resistance by repressing auxin signalling. Science 312, 436–439 (2006).
Von Bodman, S. B., Bauer, W. D. & Coplin, D. L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41, 455–482 (2003).
Leigh, J. A. & Coplin, D. L. Exopolysaccharides in plant–bacterial interactions. Annu. Rev. Microbiol. 46, 307–346 (1992).
Koutsoudis, M. D., Tsaltas, D., Minogue, T. D. & von Bodman, S. B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl Acad. Sci. USA 103, 5983–5988 (2006).
Ausubel, F. M. Are innate immune signaling pathways in plants and animals conserved? Nature Immunol. 6, 973–979 (2005).
He, P. et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK Arabidopsis innate immunity. Cell 125, 563–575 (2006). In Arabidopsis protoplasts, AvrPto and AvrPtoB suppress basal defence signalling upstream of MAPK activation.
Gomez-Gomez, L. & Boller, T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256 (2002).
Zipfel, C. & Felix, G. Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 8, 353–360 (2005).
Zeidler, D. et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl Acad. Sci. USA 101, 15811–15816 (2004).
Felix, G., Duran, J. D., Volko, S. & Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 (1999).
Kunze, G. et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507 (2004).
Felix, G. & Boller, T. Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208 (2003).
Gomez-Gomez, L. & Boller, T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 (2000).
Gomez-Gomez, L., Felix, G. & Boller, T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284 (1999).
Asai, T. et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 (2002).
Navarro, L. et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128 (2004).
Zipfel, C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004).
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T. & Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476 (2006). This paper provides biochemical evidence that the PRR FLS2 binds to flagellin.
Robatzek, S., Chinchilla, D. & Boller, T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542 (2006).
Thilmony, R., Underwood, W. & He, S. Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53 (2006).
Truman, W., de Zabala, M. T. & Grant, M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 46, 14–33 (2006). References 56 and 57 report microarray gene expression profiling of Arabidopsis that identified genes and pathways that are involved in basal defences.
Pedley, K. F. & Martin, G. B. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541–547 (2005).
Zipfel, C. et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 (2006). This paper, along with reference 47, reports that the PAMP EF-Tu is recognized by the Arabidopsis PRR EFR.
Pfund, C. et al. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17, 696–706 (2004).
Taguchi, F. et al. Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol. 44, 342–349 (2003).
Sun, W., Dunning, F. M., Pfund, C., Weingarten, R. & Bent, A. F. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18, 764–779 (2006). Variation of flagellin in Xanthomonas probably allows the pathogen to avoid PRR-mediated detection in Arabidopsis.
Jakobek, J. L., Smith, J. A. & Lindgren, P. B. Suppression of bean defense responses by Pseudomonas syringae. Plant Cell 5, 57–63 (1993).
Brown, I., Mansfield, J. W. & Bonas, U. hrp genes in Xanthomonas campestris pv. vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol. Plant Microbe Interact. 8, 825–836 (1995).
Hauck, P., Thilmony, R. & He, S. Y. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl Acad. Sci. USA 100, 8577–8582 (2003).
Li, X. et al. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl Acad. Sci. USA 102, 12990–12995 (2005).
Oh, H. S. & Collmer, A. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J. 44, 348–359 (2005).
Metz, M. et al. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 41, 801–814 (2005).
Kim, M. G. et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121, 749–759 (2005).
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R. & Dangl, J. L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389 (2003).
Mackey, D., Holt, B. F., Wiig, A. & Dangl, J. L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754 (2002).
Axtell, M. J. & Staskawicz, B. J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377 (2003).
Lim, M. T. & Kunkel, B. N. The pseudomonas syringae avrRpt2 gene contributes to virulence on tomato. Mol. Plant Microbe Interact. 18, 626–633 (2005).
Belkhadir, Y., Nimchuk, Z., Hubert, D. A., Mackey, D. & Dangl, J. L. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16, 2822–2835 (2004).
Chisholm, S. T. et al. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl Acad. Sci. USA 102, 2087–2092 (2005).
de Torres, M. et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 22 June 2006 (doi: 10.1111/j.1365–313X.2006.02798.x).
Lam, E. Controlled cell death, plant survival and development. Nature Rev. Mol. Cell Biol. 5, 305–315 (2004).
Martin, G. B., Bogdanove, A. J. & Sessa, G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61 (2003).
Tang, X. et al. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063 (1996).
Scofield, S. R. et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065 (1996).
Deslandes, L. et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl Acad. Sci. USA 100, 8024–8029 (2003).
Van der Biezen, E. A. & Jones, J. D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456 (1998).
Axtell, M. J., Chisholm, S. T., Dahlbeck, D. & Staskawicz, B. J. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49, 1537–1546 (2003).
Shan, L., He, P., Zhou, J. M. & Tang, X. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant Microbe Interact. 13, 592–598 (2000).
Pedley, K. F. & Martin, G. B. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243 (2003).
Salmeron, J. M. et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86, 123–133 (1996).
Wulf, J., Pascuzzi, P. E., Fahmy, A., Martin, G. B. & Nicholson, L. K. The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12, 1257–1268 (2004).
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Espinosa, A. & Alfano, J. R. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol. 6, 1027–1040 (2004).
Abramovitch, R. B., Kim, Y.-J., Chen, S., Dickman, M. B. & Martin, G. B. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibiton of host programmed cell death. EMBO J. 22, 60–69 (2003).
Jamir, Y. et al. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565 (2004).
Jackson, R. W. et al. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl Acad. Sci. USA 96, 10875–10880 (1999).
Tsiamis, G. et al. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19, 3204–3214 (2000).
Makino, S., Sugio, A., White, F. & Bogdanove, A. J. Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 19, 240–249 (2006).
Kim, Y. J., Lin, N. C. & Martin, G. B. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598 (2002).
Zeng, L. R. et al. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808 (2004). This paper reports the cloning of a rice U-box E3 ubiquitin ligase that regulates plant cell death and defence.
Gonzalez-Lamothe, R. et al. The u-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083 (2006).
Yang, C. W. et al. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18, 1084–1098 (2006).
Lin, N. C. & Martin, G. B. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant Microbe Interact. 18, 43–51 (2005).
Guttman, D. S. et al. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726 (2002).
Lin, N. C., Abramovitch, R. B., Kim, Y. J. & Martin, G. B. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72, 702–712 (2006).
Abramovitch, R. B. & Martin, G. B. AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245, 1–8 (2005).
Uppalapati, S. R. et al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42, 201–217 (2005).
Cui, J. et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl Acad. Sci. USA 102, 1791–1796 (2005).
Zhao, Y. et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36, 485–499 (2003).
He, P. et al. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37, 589–602 (2004).
Cohn, J. R. & Martin, G. B. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44, 139–154 (2005).
Song, W. Y. et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806 (1995).
Shimizu, R. et al. The DeltafliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol. Genet. Genomics 269, 21–30 (2003).
DebRoy, S., Thilmony, R., Kwack, Y. B., Nomura, K. & He, S. Y. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl Acad. Sci. USA 101, 9927–9932 (2004).
Mysore, K. S. et al. Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315 (2002).
Pedley, K. F. & Martin, G. B. Identification of MAPKs and their possible MAPKK activators involved in the Pto-mediated defense response of tomato. J. Biol. Chem. 279, 49229–49235 (2004).
del Pozo, O., Pedley, K. F. & Martin, G. B. MAPKKKa is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072–3082 (2004).
Schornack, S., Meyer, A., Romer, P., Jordan, T. & Lahaye, T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 163, 256–272 (2006).
Fujikawa, T., Ishihara, H., Leach, J. E. & Tsuyumu, S. Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol. Plant Microbe Interact. 19, 342–349 (2006).
Gu, K. et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125 (2005).
Anderson, J. C., Pascuzzi, P. E., Xiao, F., Sessa, G. & Martin, G. B. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18, 502–514 (2006).
Mudgett, M. B. & Staskawicz, B. J. Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32, 927–941 (1999).
Meindl, T., Boller, T. & Felix, G. The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12, 1783–1794 (2000).
Dow, M., Newman, M. A. & von Roepenack, E. The Induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261 (2000).
Acknowledgements
We thank Tracy Rosebrock and Ann Taylor for critical reading of the manuscript. We are grateful to Tom Burr, Amy Charkowski, and Paul Frey for providing photographs for Figure 1. Research in our laboratory is supported by the National Science Foundation, National Institutes of Health, United States Department of Agriculture-National Research Initiative, Binational Agriculture Research Fund, Binational Science Foundation and the Triad Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary table 1 (PDF 148 kb)
Related links
Glossary
- Stomate
-
A natural opening on leaves and stems. Stomata can open and close to ensure efficient exchange of gases and moisture in the apoplast.
- Apoplast
-
The intercellular space in the plant tissue, including the cell wall, that is outside the plasma membrane, through which nutrients and water can freely diffuse.
- Xylem
-
A network of cells in the vascular system of a plant that moves water and minerals.
- Virulence
-
Increases in the rate of growth and final population size, or enhanced disease symptoms, that promote the spread of the pathogen through the plant or in nature.
- Type III secretion system
-
A bacterial membrane-spanning protein complex, extended by a pilus. This complex functions like a syringe to inject bacterial proteins into the host cell cytoplasm.
- Effector
-
A bacterial protein that is translocated by the type III secretion system into the plant cell cytoplasm.
- Pathogen-associated molecular patterns
-
(PAMPs). Bacterial molecules that have an important role in the microbial lifestyle, and that contain a conserved feature that is recognized by a pathogen recognition receptor (PRR).
- Pathogen recognition receptor
-
(PRR). A host receptor, such as FLS2 or EFR, that can detect the presence of pathogens by recognizing conserved pathogen molecules (such as PAMPs).
- Basal defence
-
Plant defence that occurs early in the host–pathogen interaction in response to the perception by plant pattern recognition receptors (PRRs) of extracellular pathogen-associated molecular patterns (PAMPs).
- Biotrophy
-
A period of colonization during which a microorganism relies on living host tissue to grow.
- Hormone
-
A signal molecule that is produced at specific locations and at low concentrations. Hormones can be transported throughout the plant and regulate biological processes.
- Arabidopsis
-
A plant of the mustard family that is used as a model organism in plant molecular biology.
- Callose
-
A polysaccharide that is a common plant cell wall constituent and that is deposited near infection sites in structures known as papillae. Callose deposition is associated with basal defences and is believed to limit pathogen virulence.
- Resistance (R) protein
-
A plant protein that recognizes, either directly or indirectly, a specific pathogen avirulence protein (often a type III effector) to activate plant immunity.
- Gene-for-gene model of disease resistance
-
A model for plant immunity in which plant resistance genes are only effective if a specific avirulence gene is expressed by the pathogen.
- Avirulence (Avr) protein
-
A pathogen protein that elicits plant immunity in plants that express a specific resistance protein. Avr proteins are often type III effector proteins.
- Hypersensitive response
-
(HR). A defence that is often associated with resistance (R)-protein-mediated immunity. During the HR, the plant initiates programmed cell death in cells that surround the pathogen to inhibit pathogen spreading.
Rights and permissions
About this article
Cite this article
Abramovitch, R., Anderson, J. & Martin, G. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7, 601–611 (2006). https://doi.org/10.1038/nrm1984
Issue Date:
DOI: https://doi.org/10.1038/nrm1984
This article is cited by
-
iTRAQ based proteomic analysis of rice lines having single or stacked blast resistance genes: Pi54/Pi54rh during incompatible interaction with Magnaporthe oryzae
Physiology and Molecular Biology of Plants (2023)
-
Heterologous production and characterization of a pyomelanin of Antarctic Pseudomonas sp. ANT_H4: a metabolite protecting against UV and free radicals, interacting with iron from minerals and exhibiting priming properties toward plant hairy roots
Microbial Cell Factories (2022)
-
Characterization of the belowground microbial community and co-occurrence networks of tobacco plants infected with bacterial wilt disease
World Journal of Microbiology and Biotechnology (2022)
-
Altered metabolomic states elicited by Flg22 and FlgII-28 in Solanum lycopersicum: intracellular perturbations and metabolite defenses
BMC Plant Biology (2021)
-
Unveiling the putative functional genes present in root-associated endophytic microbiome from maize plant using the shotgun approach
Journal of Applied Genetics (2021)