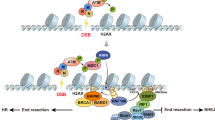
Key Points
-
The repair of chromosomal DNA double-strand breaks, which is essential for the maintenance of genomic stability, occurs within the context of chromatin. Histone modifications correlate with DNA damage and might therefore serve as a code for repair.
-
Chromatin-remodelling complexes and histone-modifying enzymes are recruited to the sites of DNA damage. Histone tails and core domains also show significant damage-correlated modifications, including acetylation, deacetylation, methylation, phosphorylation and ubiquitylation.
-
Histone H2A phosphorylation by the ATM/ATR kinase has a key role in the recruitment of the INO80 chromatin remodeller and in the loading of cohesin at sites of double-strand breaks. Cohesin loading near DNA double-strand breaks might be facilitated by both the Mre11 protein and its ligand, the chromatin remodelling complex RSC.
-
A histone remodeler related to INO80, the SWR1 complex, can exchange histone H2A for a variant known as Htz1. Genetics studies implicate SWR1 in DNA repair, and the analysis of a related complex in Drosophila, TIP60, indicates that TIP60 might replace phospho-H2Av with unmodified histone at sites of damage.
-
Histone acetylation and deacetylation by the NuA4 and Sin3–Rpd3 complexes, respectively, might help open compact nucleosomal fibers at the sites of DNA damage. Roles for other covalent modifications of histones in facilitating repair are suggested but not definitively proven.
-
Histone H3K79 and H4K20 methylation is important to allow the recruitment of the checkpoint adaptor proteins 53BP1 (mammals) and Crb2 (fission yeast), respectively, to the sites of damage. This links histone modification with checkpoint activation, as the adaptors seem to bind at the sites of damage to stimulate checkpoint effector kinase activation.
Abstract
Chromatin modifications are important for all cellular processes that involve DNA, including transcription, replication and DNA repair. Chromatin can be modified by the addition of adducts to histone tail residues or by nucleosome remodelling, which requires ATP-dependent chromatin-remodelling complexes. Although the role of these mechanisms in transcription is well studied, their impact on DNA repair has only recently become evident. One crucial chromatin modification, the phosphorylation of histone H2A, links the recruitment of histone modifiers and ATP-dependent chromatin-remodelling complexes to sites of DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
van Gent, D. C., Hoeijmakers, J. H. & Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet. 2, 196–206 (2001).
Marmorstein, R. Protein modules that manipulate histone tails for chromatin regulation. Nature Rev. Mol. Cell Biol. 2, 422–432 (2001).
Lusser, A. & Kadonaga, J. T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25, 1192–1200 (2003).
Stiff, T. et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64, 2390–2396 (2004).
Burma, S., Chen, B. P., Murphy, M., Kurimasa, A. & Chen, D. J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467 (2001).
Ward, I. M. & Chen, J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Downs, J. A., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Shroff, R. et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14, 1703–1711 (2004). Shows that the checkpoint kinases Mec1 and Tel1 are responsible for the phosphorylation of histone H2A within a ∼50-kb region near DSBs.
Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K. & Sugimoto, K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294, 867–870 (2001).
Melo, J. A., Cohen, J. & Toczyski, D. P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809–2821 (2001).
Nakada, D., Matsumoto, K. & Sugimoto, K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17, 1957–1962 (2003).
Rouse, J. & Jackson, S. P. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9, 857–869 (2002).
Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97, 621–633 (1999).
Wolner, B., van Komen, S., Sung, P. & Peterson, C. L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12, 221–232 (2003).
Sugawara, N., Wang, X. & Haber, J. E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12, 209–219 (2003).
Redon, C. et al. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4, 678–684 (2003).
Wyatt, H. R., Liaw, H., Green, G. R. & Lustig, A. J. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164, 47–64 (2003).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Bassing, C. H. et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl Acad. Sci. USA 99, 8173–8178 (2002).
Bassing, C. H. et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114, 359–70 (2003).
Celeste, A. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003).
Xie, A. et al. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16, 1017–1025 (2004).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003).
Nakamura, T. M., Du, L. L., Redon, C. & Russell, P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell Biol. 24, 6215–6230 (2004).
Krogan, N. J. et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576 (2003). This report, together with references 28 and 29, describes the purification of the SWR1 chromatin-remodelling complex that facilitates the incorporation of the Htz1 histone variant in chromatin.
Shen, X., Mizuguchi, G., Hamiche, A. & Wu, C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Kobor, M. S. et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131 (2004).
Downs, J. A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004). Describes the interaction between phospho-H2A peptides and Arp4, a member of the NuA4, INO80 and SWR1 chromatin-modifying complexes, which seems to be important for the recruitment of these Arp4-containing complexes to phosphorylated H2A near the sites of damage.
Bird, A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415 (2002).
Choy, J. S. & Kron, S. J. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell Biol. 22, 8215–8225 (2002).
Jazayeri, A., McAinsh, A. D. & Jackson, S. P. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl Acad. Sci. USA 101, 1644–1649 (2004).
Kurdistani, S. K. & Grunstein, M. Histone acetylation and deacetylation in yeast. Nature Rev. Mol. Cell Biol. 4, 276–284 (2003).
Tsaneva, I. R., Muller, B. & West, S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69, 1171–1180 (1992).
van Attikum, H., Fritsch, O., Hohn, B. & Gasser, S. M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 (2004).
Morrison, A. J. et al. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 (2004). This report, as well as references 30 and 36, shows the recruitment of the INO80 chromatin-remodelling complex to DSBs through interaction with phosphorylated histone H2A, and that recruitment requires the INO80 subunit Nhp10.
Fritsch, O., Benvenuto, G., Bowler, C., Molinier, J. & Hohn, B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16, 479–485 (2004). Reports the first isolation of an orthologue of the yeast Ino80 SWI2/SNF2 ATPase in a higher eukaryote, Arabidopsis thaliana , and shows a function for A. thaliana INO80 in the homologous recombination pathway for DNA damage repair.
Ikura, T. et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000).
Doyon, Y. & Cote, J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14, 147–154 (2004).
Madigan, J. P., Chotkowski, H. L. & Glaser, R. L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30, 3698–3705 (2002).
Kusch, T. et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087 (2004). Reports a functional analysis of the D. melanogaster TIP60 complex, which led to the finding that D. melanogaster TIP60 acetylates phosphorylated H2Av, a D. melanogaster -specific histone variant, and exchanges it with unmodified H2Av in response to DNA damage.
Shim, E. Y., Ma, J. L., Oum, J. H., Yanez, Y. & Lee, S. E. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell Biol. 25, 3934–3944 (2005).
Bennett, C. B. et al. Genes required for ionizing radiation resistance in yeast. Nature Genet. 29, 426–434 (2001).
Koyama, H., Itoh, M., Miyahara, K. & Tsuchiya, E. Abundance of the RSC nucleosome-remodeling complex is important for the cells to tolerate DNA damage in Saccharomyces cerevisiae. FEBS Lett. 531, 215–221 (2002).
Chai, B., Huang, J., Cairns, B. R. & Laurent, B. C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661 (2005).
Angus-Hill, M. L. et al. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7, 741–751 (2001).
Sudarsanam, P., Iyer, V. R., Brown, P. O. & Winston, F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 97, 3364–3369 (2000).
Sjogren, C. & Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 (2001).
Kim, J. S., Krasieva, T. B., LaMorte, V., Taylor, A. M. & Yokomori, K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277, 45149–45153 (2002).
Unal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
Strom, L., Lindroos, H. B., Shirahige, K. & Sjogren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004). References 51 and 52 show the de novo recruitment of cohesin to DSBs, which requires phosphorylated histone H2A and the Mre11 protein, and facilitates postreplicational repair.
Huang, J., Hsu, J. M. & Laurent, B. C. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms. Mol. Cell 13, 739–50 (2004).
Harvey, A. C., Jackson, S. P. & Downs, J. A. Saccharomyces cerevisiae histone H2A Ser122 facilitates DNA repair. Genetics 170, 543–553 (2005).
Fernandez-Capetillo, O., Allis, C. D. & Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 199, 1671–1677 (2004).
Cheung, W. L. et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507–517 (2003).
Ahn, S. H. et al. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120, 25–36 (2005).
Giannattasio, M., Lazzaro, F., Plevani, P. & Muzi-Falconi, M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6–Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280, 9879–9886 (2005).
Briggs, S. D. et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418, 498 (2002).
Singer, M. S. et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150, 613–632 (1998).
Huyen, Y. et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 (2004).
Sanders, S. L. et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 (2004).
Qin, S. & Parthun, M. R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell Biol. 22, 8353–8365 (2002).
Cheung, W. L. et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15, 656–660 (2005).
Acknowledgements
The authors acknowledge the European Molecular Biology Organization (EMBO) and the International Human Frontier Science Program (HFSP) Organization for fellowships to H.v.A., and thank the Swiss Cancer League, the European RTN Checkpoints and Cancer, the Swiss National Science Foundation and the Novartis Research Foundation for their support. The authors also thank Brehon Laurent for sharing results before publication and apologize to all researchers whose work could not be discussed due to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Saccharomyces Genome Database
Swiss-Prot
FURTHER INFORMATION
Glossary
- CHROMATIN
-
A higher-order structure of DNA folded around histone octamers and stabilized by linker histones and other factors.
- NUCLEOSOME
-
The basic unit of chromatin composed of 147 bp of chromosomal DNA wrapped around an octamer that contains two copies of each histone H2A, H2B, H3 and H4, or appropriate histone variants.
- CHROMATIN IMMUNOPRECIPITATION
-
(ChIP). A technique that allows the study of protein–DNA interactions by the amplification of DNA sequences from complexes of crosslinked proteins and DNA, recovered by immunoprecipitation with antibodies against the proteins in question.
- BROMODOMAIN
-
An evolutionary conserved protein domain that can bind to acetylated residues of histones.
- SANT DOMAIN
-
An evolutionary conserved protein domain that is important for DNA and histone-tail binding.
- CHROMODOMAIN
-
An evolutionary conserved protein domain that can bind to methylated residues of histones.
- EUCHROMATIN
-
Decondensed regions of chromatin usually associated with active transcription.
- HISTONE ACETYLTRANSFERASE
-
(HAT). An enzyme that adds acetyl groups to lysine or arginine residues of a histone.
- HISTONE DEACETYLASE
-
(HDAC). An enzyme that removes acetyl groups from lysine or arginine residues of a histone.
- HETEROCHROMATIN
-
Condensed regions of chromatin usually associated with the repression of transcription and late replication.
- HISTONE METHYLTRANSFERASE
-
An enzyme that adds methyl groups to lysine or arginine residues of histones.
- TUDOR DOMAIN
-
An evolutionary conserved chromodomain-like protein domain that can bind to methylated residues of histones.
Rights and permissions
About this article
Cite this article
van Attikum, H., Gasser, S. The histone code at DNA breaks: a guide to repair?. Nat Rev Mol Cell Biol 6, 757–765 (2005). https://doi.org/10.1038/nrm1737
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1737
This article is cited by
-
COMMD4 functions with the histone H2A-H2B dimer for the timely repair of DNA double-strand breaks
Communications Biology (2021)
-
Elevated HDAC activity and altered histone phospho-acetylation confer acquired radio-resistant phenotype to breast cancer cells
Clinical Epigenetics (2020)
-
Chromatin states responsible for the regulation of differentially expressed genes under 60Co~γ ray radiation in rice
BMC Genomics (2017)
-
The Bucentaur (BCNT) protein family: a long-neglected class of essential proteins required for chromatin/chromosome organization and function
Chromosoma (2015)
-
A gene ontology inferred from molecular networks
Nature Biotechnology (2013)