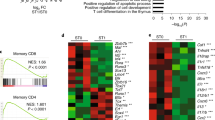
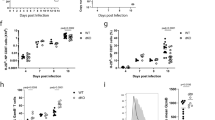
Key Points
-
BTB and CNC homology (BACH) proteins are transcriptional repressors of the basic region leucine zipper (bZIP) transcription factor family.
-
BACH proteins have widespread roles in immunological processes, including tolerance, memory, immunosuppression and iron homeostasis.
-
BACH proteins stabilize lineage commitment and promote cell-type-specific functions through the repression of alternative lineage programmes.
-
Competitive interactions with transcriptional activators of the bZIP family form a common mechanistic theme underlying the diverse functions of BACH factors.
-
The expression and function of BACH factors are regulated by extrinsic signals to enable fine-tuning of cellular functions.
Abstract
BTB and CNC homology (BACH) proteins are transcriptional repressors of the basic region leucine zipper (bZIP) transcription factor family. Recent studies indicate widespread roles of BACH proteins in controlling the development and function of the innate and adaptive immune systems, including the differentiation of effector and memory cells of the B and T cell lineages, CD4+ regulatory T cells and macrophages. Here, we emphasize similarities at a molecular level in the cell-type-specific activities of BACH factors, proposing that competitive interactions of BACH proteins with transcriptional activators of the bZIP family form a common mechanistic theme underlying their diverse actions. The findings contribute to a general understanding of how transcriptional repressors shape lineage commitment and cell-type-specific functions through repression of alternative lineage programmes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rothenberg, E. V. & Scripture-Adams, D. D. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin. Immunol. 20, 236–246 (2008).
Huang, S., Guo, Y. P., May, G. & Enver, T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713 (2007).
Oyake, T. et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095 (1996). The discovery and initial characterization of BACH1 and BACH2 as MAF partners.
Reinke, A. W., Baek, J., Ashenberg, O. & Keating, A. E. Networks of bZIP protein–protein interactions diversified over a billion years of evolution. Science 340, 730–734 (2013).
Turner, R. & Tjian, R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos–cJun heterodimers. Science 243, 1689–1694 (1989).
Glover, J. N. & Harrison, S. C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature 373, 257–261 (1995). The crystal structure of FOS–JUN AP-1 dimers at DNA.
Hai, T. & Curran, T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl Acad. Sci. USA 88, 3720–3724 (1991).
Kuwahara, M. et al. Bach2–Batf interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat. Commun. 7, 12596 (2016). The identification of competitive interactions between BACH2–BATF repressor complexes and JUND–BATF–IRF4 activator complexes at a type 2 cytokine locus control region.
Roychoudhuri, R. et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016). The discovery of the function of BACH2 in memory CD8+ T cell differentiation and competitive interactions of BACH2 with JUN family AP-1 factors in lymphocytes.
Jindrich, K. & Degnan, B. M. The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol. 16, 28 (2016).
Amoutzias, G. D. et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 24, 827–835 (2007).
Smith, J. J. et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45, 415–421 (2013).
Herrin, B. R. & Cooper, M. D. Alternative adaptive immunity in jawless vertebrates. J. Immunol. 185, 1367–1374 (2010).
Escriva, H., Manzon, L., Youson, J. & Laudet, V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol. Biol. Evol. 19, 1440–1450 (2002).
Itoh-Nakadai, A. et al. The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nat. Immunol. 15, 1171–1180 (2014). The discovery of the functions of BACH2 and BACH1 in promoting B cell differentiation from common lymphoid progenitors.
Chaharbakhshi, E. & Jemc, J. C. Broad-complex, tramtrack, and bric-a-brac (BTB) proteins: critical regulators of development. Genesis 54, 505–518 (2016).
Igarashi, K. & Watanabe-Matsui, M. Wearing red for signaling: the heme–bach axis in heme metabolism, oxidative stress response and iron immunology. Tohoku J. Exp. Med. 232, 229–253 (2014).
Igarashi, K. et al. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J. Biol. Chem. 273, 11783–11790 (1998).
Yoshida, C. et al. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4, 643–655 (1999).
Tanaka, H. et al. Epigenetic regulation of the Blimp-1 Gene (Prdm1) in B cells involves Bach2 and histone deacetylase 3. J. Biol. Chem. 291, 6316–6330 (2016).
Hoshino, H. et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J. Biol. Chem. 275, 15370–15376 (2000).
Hira, S., Tomita, T., Matsui, T., Igarashi, K. & Ikeda-Saito, M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life 59, 542–551 (2007).
Watanabe-Matsui, M. et al. Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood 117, 5438–5448 (2011). The identification of haem as a regulator of BACH2.
Watanabe-Matsui, M. et al. Heme binds to an intrinsically disordered region of Bach2 and alters its conformation. Arch. Biochem. Biophys. 565, 25–31 (2015).
Tompa, P., Schad, E., Tantos, A. & Kalmar, L. Intrinsically disordered proteins: emerging interaction specialists. Curr. Opin. Struct. Biol. 35, 49–59 (2015).
Suenaga, T. et al. Charge-state-distribution analysis of Bach2 intrinsically disordered heme binding region. J. Biochem. 160, 291–298 (2016).
Haldar, M. et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014). The discovery of the function of BACH1 in regulating bone marrow and red pulp macrophage differentiation.
Sun, J. et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21, 5216–5224 (2002).
Brand, M. et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 11, 73–80 (2004).
Ogawa, K. et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 (2001).
Suzuki, H. et al. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 23, 2544–2553 (2004).
Zenke-Kawasaki, Y. et al. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 27, 6962–6971 (2007).
Alam, J. et al. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am. J. Physiol. Renal Physiol. 284, F743–F752 (2003).
Macian, F., Lopez-Rodriguez, C. & Rao, A. Partners in transcription: NFAT and AP-1. Oncogene 20, 2476–2489 (2001).
Murphy, T. L., Tussiwand, R. & Murphy, K. M. Specificity through cooperation: BATF–IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13, 499–509 (2013).
Muto, A. et al. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J. Biol. Chem. 277, 20724–20733 (2002).
Lee, G. R., Fields, P. E., Griffin, T. J. & Flavell, R. A. Regulation of the Th2 cytokine locus by a locus control region. Immunity 19, 145–153 (2003).
Hama, M. et al. Bach1 regulates osteoclastogenesis in a mouse model via both heme oxygenase 1-dependent and heme oxygenase 1-independent pathways. Arthritis Rheum. 64, 1518–1528 (2012).
Bhandoola, A. & Sambandam, A. From stem cell to T cell: one route or many? Nat. Rev. Immunol. 6, 117–126 (2006).
Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 238, 37–46 (2010).
Pongubala, J. M. et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9, 203–215 (2008).
Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Ganz, T. Macrophages and systemic iron homeostasis. J. Innate Immun. 4, 446–453 (2012).
Sun, J. et al. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl Acad. Sci. USA 101, 1461–1466 (2004).
Tan, M. K., Lim, H. J., Bennett, E. J., Shi, Y. & Harper, J. W. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24 (2013).
Gottwein, E. et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099 (2007).
Skalsky, R. L. et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8, e1002484 (2012).
Ota, K., Brydun, A., Itoh-Nakadai, A., Sun, J. & Igarashi, K. Bach1 deficiency and accompanying overexpression of heme oxygenase-1 do not influence aging or tumorigenesis in mice. Oxid. Med. Cell. Longev. 2014, 757901 (2014).
Yano, Y. et al. Genetic ablation of the transcription repressor Bach1 leads to myocardial protection against ischemia/reperfusion in mice. Genes Cells 11, 791–803 (2006).
Kanno, H. et al. Genetic ablation of transcription repressor Bach1 reduces neural tissue damage and improves locomotor function after spinal cord injury in mice. J. Neurotrauma 26, 31–39 (2009).
Yamada, K. et al. Modulation of the secondary injury process after spinal cord injury in Bach1-deficient mice by heme oxygenase-1. J. Neurosurg. Spine 9, 611–620 (2008).
Iida, A. et al. Bach1 deficiency ameliorates hepatic injury in a mouse model. Tohoku J. Exp. Med. 217, 223–229 (2009).
Tanimoto, T. et al. Genetic ablation of the Bach1 gene reduces hyperoxic lung injury in mice: role of IL-6. Free Radic. Biol. Med. 46, 1119–1126 (2009).
Watari, Y. et al. Ablation of the bach1 gene leads to the suppression of atherosclerosis in bach1 and apolipoprotein E double knockout mice. Hypertens.Res. 31, 783–792 (2008).
Harusato, A. et al. BTB and CNC homolog 1 (Bach1) deficiency ameliorates TNBS colitis in mice: role of M2 macrophages and heme oxygenase-1. Inflamm. Bowel Dis. 19, 740–753 (2013).
Martinez, F. O. & Gordon, S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 6, 13 (2014).
So, A. Y. et al. Regulation of APC development, immune response, and autoimmunity by Bach1/HO-1 pathway in mice. Blood 120, 2428–2437 (2012).
Naito, Y., Takagi, T. & Higashimura, Y. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch. Biochem. Biophys. 564, 83–88 (2014).
Kanzaki, H. et al. RANKL induces Bach1 nuclear import and attenuates Nrf2-mediated antioxidant enzymes, thereby augmenting intracellular reactive oxygen species signaling and osteoclastogenesis in mice. FASEB J. 31, 781–792 (2017).
Roychoudhuri, R. et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498, 506–510 (2013). The discovery of the role of BACH2 in immunological tolerance and in the regulation of CD4+ T reg cell and effector T cell differentiation.
Kim, E. H. et al. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J. Immunol. 192, 985–995 (2014).
Nakamura, A. et al. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J. Exp. Med. 210, 2191–2204 (2013). The identification and characterization of pulmonary alveolar proteinosis in Bach2 -deficient mice.
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4, 337–342 (2003).
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). References 63–65 describe how the transcription factor FOXP3 fulfils a crucial function in immunological tolerance by promoting the differentiation of CD4+ T reg cells.
Ohkura, N., Kitagawa, Y. & Sakaguchi, S. Development and maintenance of regulatory T cells. Immunity 38, 414–4232 (2013).
Tang, Q. & Bluestone, J. A. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat. Immunol. 9, 239–244 (2008).
Josefowicz, S. Z., Lu, L. F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Quezada, S. A., Peggs, K. S., Simpson, T. R. & Allison, J. P. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol. Rev. 241, 104–118 (2011).
Roychoudhuri, R., Eil, R. L. & Restifo, N. P. The interplay of effector and regulatory T cells in cancer. Curr. Opin. Immunol. 33, 101–111 (2015).
Roychoudhuri, R. et al. The transcription factor BACH2 promotes tumor immunosuppression. J. Clin. Invest. 126, 599–604 (2016). The discovery of the function of BACH2 in promoting tumour immunosuppression.
van der Weyden, L. et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature 541, 233–236 (2017).
Ascierto, M. L. et al. The intratumoral balance between metabolic and immunologic gene expression is associated with anti-PD-1 response in patients with renal cell carcinoma. Cancer Immunol. Res. 4, 726–733 (2016).
Tsukumo, S. et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc. Natl Acad. Sci. USA 110, 10735–10740 (2013).
Kuwahara, M. et al. The Menin–Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat. Commun. 5, 3555 (2014). The discovery of the function of BACH2 in suppressing cellular senescence in CD4+ T cells.
Restifo, N. P. & Gattinoni, L. Lineage relationship of effector and memory T cells. Curr. Opin. Immunol. 25, 556–563 (2013).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Belz, G. T. & Kallies, A. Effector and memory CD8+ T cell differentiation: toward a molecular understanding of fate determination. Curr. Opin. Immunol. 22, 279–285 (2010).
Hedrick, S. M., Hess Michelini, R., Doedens, A. L., Goldrath, A. W. & Stone, E. L. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12, 649–661 (2012).
Chang, J. T., Wherry, E. J. & Goldrath, A. W. Molecularregulation of effector and memory T cell differentiation. Nat. Immunol. 15, 1104–1115 (2014).
Teixeiro, E. et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science 323, 502–505 (2009).
Wirth, T. C. et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity 33, 128–140 (2010).
Roychoudhuri, R. et al. Transcriptional profiles reveal a stepwise developmental program of memory CD8+ T cell differentiation. Vaccine 33, 914–923 (2015).
Hu, G. & Chen, J. A genome-wide regulatory network identifies key transcription factors for memory CD8+ T-cell development. Nat. Commun. 4, 2830 (2013).
Kaech, S. M. et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). The identification of memory precursor effector cells as progenitors of CD8+ T cell memory.
Cui, W. & Kaech, S. M. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol. Rev. 236, 151–166 (2010).
Opferman, J. T. et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426, 671–676 (2003).
Ikeda, T., Shibata, J., Yoshimura, K., Koito, A. & Matsushita, S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 195, 716–725 (2007).
Maldarelli, F. et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). References 88 and 89 describe recurrent HIV-1 proviral integration in the BACH2 locus of CD4+ T cells that are persistently infected after initiation of ART.
Wagner, T. A. et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014).
Kurosaki, T., Kometani, K. & Ise, W. Memory B cells. Nat. Rev. Immunol. 15, 149–159 (2015).
Muto, A. et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429, 566–571 (2004). The discovery of the requirement for BACH2 in CSR and SHM of immunoglobulin genes.
Shinnakasu, R. et al. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17, 861–869 (2016). This paper identifies that BACH2 expression levels have an instructive role in the differentiation of memory B cells in the germinal centre.
Ochiai, K. et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J. Biol. Chem. 281, 38226–38234 (2006).
Muto, A. et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 29, 4048–4061 (2010). A demonstration of the requirement of BACH2-mediated Prdm1 repression in CSR of immunoglobulin genes.
Kometani, K. et al. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity 39, 136–147 (2013). A demonstration that expression levels of BACH2 are a crucial determinant of the functional activity of memory B cells.
Ochiai, K., Muto, A., Tanaka, H., Takahashi, S. & Igarashi, K. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int. Immunol. 20, 453–460 (2008).
Huang, C., Geng, H., Boss, I., Wang, L. & Melnick, A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood 123, 1012–1020 (2014).
Smith, K. G., Light, A., Nossal, G. J. & Tarlinton, D. M. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 16, 2996–3006 (1997).
Zotos, D. et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207, 365–378 (2010).
Linterman, M. A. et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207, 353–363 (2010).
Ochiai, K. et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38, 918–929 (2013).
Heise, N. et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J. Exp. Med. 211, 2103–2118 (2014).
Willinger, T., Freeman, T., Hasegawa, H., McMichael, A. J. & Callan, M. F. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J. Immunol. 175, 5895–5903 (2005).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Jang, K. J. et al. Mitochondrial function provides instructive signals for activation-induced B-cell fates. Nat. Commun. 6, 6750 (2015). A demonstration of the regulation of plasma cell differentiation by haem metabolism.
Ando, R. et al. The transcription factor Bach2 is phosphorylated at multiple sites in murine B cells but a single site prevents its nuclear localization. J. Biol. Chem. 291, 1826–1840 (2016). The identification of BACH2 phosphorylation sites in B cells using mass spectrometry.
Yoshida, C. et al. Bcr–Abl signaling through the PI-3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase-1. Blood 109, 1211–1219 (2007).
Heng, T. S., Painter, M. W. & Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Itoh-Nakadai, A. et al. Bach2-Cebp gene regulatory network for the commitment of multipotent hematopoietic progenitors. Cell Rep. 18, 2401–2414 (2017).
Zhang, D. X. & Glass, C. K. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J. Mol. Endocrinol. 51, T37–T50 (2013).
International Multiple Sclerosis Genetics Consortium. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Cooper, J. D. et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40, 1399–1401 (2008).
Dubois, P. C. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Ferreira, M. A. et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 (2011).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Jin, Y. et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680 (2012).
Reich, D. E. et al. Linkage disequilibrium in the human genome. Nature 411, 199–204 (2001).
Christodoulou, K. et al. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut 62, 977–984 (2013).
Medici, M. et al. Identification of novel genetic Loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet. 10, e1004123 (2014). References 112–120 identify polymorphic genetic variations in the BACH2 locus that are associated with immune-mediated diseases.
Swaminathan, S. et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat. Med. 19, 1014–1022 (2013).
Casolari, D. A. et al. Transcriptional suppression of BACH2 by the Bcr-Abl oncoprotein is mediated by PAX5. Leukemia 27, 409–415 (2013).
Scholtysik, R. et al. Characterization of genomic imbalances in diffuse large B-cell lymphoma by detailed SNP-chip analysis. Int. J. Cancer 136, 1033–1042 (2015).
Ichikawa, S. et al. Association between BACH2 expression and clinical prognosis in diffuse large B-cell lymphoma. Cancer Sci. 105, 437–444 (2014).
Sakane-Ishikawa, E. et al. Prognostic significance of BACH2 expression in diffuse large B-cell lymphoma: a study of the Osaka Lymphoma Study Group. J. Clin. Oncol. 23, 8012–8017 (2005).
Vassiliou, G. S. et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 43, 470–475 (2011).
Acknowledgements
R.R is supported by the Wellcome Trust−Royal Society (grant 105663/Z/14/Z), the UK Biotechnology and Biological Sciences Research Council (grant BB/N007794/1) and Cancer Research UK (grant C52623/A22597). K.I. is supported by grants-in-aid (16K15227, 15H02506, 23116003) from the Japan Society for the Promotion of Science, and AMED-CREST and P-CREATE from the Japan Agency for Medical Research and Development. T.K. is supported by grants-in-aid (A212290070, A262213060) from the Japan Society for the Promotion of Science, and CREST (J098501018) from Japan Science Technology. The authors thank N. P. Restifo, L. Gattinoni, M. Stammers, A. Muto, K. Ochiai, A. Itoh-Nakadai, M. Watanabe-Matsui, D. Clever, R. Eil, F. M. Grant, R. Nasrallah, F. Sadiyah, T. M. Lozano, K. Okkenhaug, M. Turner and G. W. Butcher for ideas and discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- TPA response elements
-
(TREs). DNA sequences (with similarity to 5′-TGA(G/C)TCA-3′) that promote gene expression induced by the phorbol ester 12-O-tetradecanoylphorbol- 13-acetate (TPA) and that are the canonical recognition sequence of the activator protein 1 (AP-1) transcription factor complexes formed by dimers of JUN and FOS.
- cAMP response elements
-
(CREs). DNA sequences (with similarity to 5′-TGACGTCA-3′) that promote gene expression induced by the cyclic AMP (cAMP) pathway and that are the canonical recognition sequence of CRE-binding protein (CREB) complexes.
- Enhancers
-
Regulatory elements that function together with promoters to control gene expression. Enhancers can lie within intronic and intergenic regions and form looping interactions to bring enhancer-bound transcription factor complexes into contact with general transcription factors assembled at promoters. Distinct repertoires of enhancers function in different cell types, allowing for cell-type-specific regulation of gene expression.
- Genetic polymorphisms
-
Genetic variations occurring in a specific population at such a frequency that the rarest of them cannot be maintained by recurrent mutation or immigration alone: therefore, they most frequently occur through inheritance.
- Thymus-derived Treg cells
-
(tTreg cells). Most of these cells develop at the CD4+ single-positive stage of thymic T cell maturation as a result of the intermediate affinity of their T cell receptors for self-antigens. These cells contribute to peripheral tolerance against self-antigens.
- Peripherally derived Treg cells
-
(pTreg cells). These cells develop from mature CD4+FOXP3− T cells in peripheral tissues. pTreg cells are frequently induced at mucosal sites, such as the lungs and gut, where they contribute to tolerance against innocuous foreign antigens.
- Progressive differentiation model
-
Proposes that naive T cells differentiate into a heterogeneous pool of memory precursor and effector T cells early during infection. Effector cells are short-lived and die following the withdrawal of antigen. Memory precursor cells survive after contraction of the effector population and give rise to memory cells. The alternative linear differentiation model proposes that naive CD8+ T cells differentiate uniformly into effector cells early during immune responses, and a subset of these differentiate into memory cells on the withdrawal of antigen.
- Cellular senescence
-
A form of irreversible growth arrest that limits the replicative lifespan of cells. Replicative senescence is induced by telomere shortening that occurs as a result of DNA replication during mitosis. Premature cellular senescence occurs in the absence of telomere shortening and results in growth arrest and apoptosis through the action of gene products of the INK4/ARF locus.
Rights and permissions
About this article
Cite this article
Igarashi, K., Kurosaki, T. & Roychoudhuri, R. BACH transcription factors in innate and adaptive immunity. Nat Rev Immunol 17, 437–450 (2017). https://doi.org/10.1038/nri.2017.26
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2017.26
This article is cited by
-
Transcription factor BACH1 in cancer: roles, mechanisms, and prospects for targeted therapy
Biomarker Research (2024)
-
BACH2-mediated CD28 and CD40LG axes contribute to pathogenesis and progression of T-cell lymphoblastic leukemia
Cell Death & Disease (2024)
-
The dual role and mutual dependence of heme/HO-1/Bach1 axis in the carcinogenic and anti-carcinogenic intersection
Journal of Cancer Research and Clinical Oncology (2023)
-
Genetics of ANCA-associated vasculitis: role in pathogenesis, classification and management
Nature Reviews Rheumatology (2022)
-
Single-cell ATAC-seq maps the comprehensive and dynamic chromatin accessibility landscape of CAR-T cell dysfunction
Leukemia (2022)