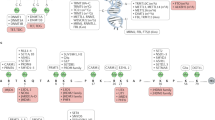
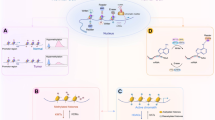
Key Points
-
Alterations in epigenetic marks — specifically DNA methylation — are an emerging biomarker that may be used in the decision-making process for disease diagnosis, prognosis and treatment, most notably in cancer, but there are examples in other diseases, such as type 1 diabetes.
-
Alterations in epigenetic marks that are chosen as biomarkers must be carefully selected owing to the dynamic nature of these marks. These alterations may be detected in non-invasive tissues, such as serum, in addition to primary tissues and so may have advantages over genetic biomarkers.
-
Glutathione S-transferase pi 1 (GSTP1) is the best-studied example of an epigenetic biomarker for cancer diagnosis. However, additional candidates show a high potential for future clinical applications, although specificity of diagnosis is an issue, and combinatorial approaches analysing DNA methylation alterations at several genes may improve this.
-
Single-gene approaches have identified epigenetic biomarkers that predict cancer recurrence and survival. Recently, high-resolution genome-wide technologies have shown the potential to improve this strategy with DNA methylation signatures of cancers showing high predictive capacity of disease prognosis.
-
DNA methylation alterations may be used as biomarkers to predict response to chemotherapy strategies. O6-methylguanine DNA methyltransferase (MGMT) and breast cancer 1, early onset (BRCA1) are examples of hypermethylated genes that predict a response to chemotherapy in cancer. Additional prognostic gene markers have already been identified and suggest DNA methylation profiling as a potent strategy to predict drug response.
-
Recent advances in sequencing and array technologies, which are capable of screening DNA methylomes genome-wide at high-resolution, gave unexpected novel insights in cancer biology. They will be crucial for DNA methylation profiling and the identification of epigenetic biomarker for diagnosis, prognosis and prediction of drug response.
Abstract
Knowledge of epigenetic alterations in disease is rapidly increasing owing to the development of genome-wide techniques for their identification. The ever-growing number of genes that show epigenetic alterations in disease emphasizes the crucial role of these epigenetic alterations — particularly DNA methylation — for future diagnosis, prognosis and prediction of response to therapies. This Review focuses on epigenetic profiling, which has started to be of clinical value in cancer and may in the future be extended to other diseases, such as neurological and autoimmune disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sandoval, J. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702 (2011).
Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Harris, R. A. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nature Biotech. 28, 1097–1105 (2010).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nature Rev. Cancer 11, 726–734 (2011).
Esteller, M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 343, 1350–1354 (2000).
Esteller, M. et al. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 58, 4515–4518 (1998).
Hon, G. C. et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2011).
Berman, B. P. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genet. 44, 40–46 (2011).
Hansen, K. D. Increased methylation variation in epigenetic domains across cancer types. Nature Genet. 43, 768–775 (2011).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Rev. Genet. 13, 484–492 (2012).
Rodríguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nature Med. 17, 330–339 (2011).
Dawson, M. A. & Kouzarides, T. Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012).
Adams, D. et al. BLUEPRINT to decode the epigenetic signature written in blood. Nature Biotech. 30, 224–226 (2012).
Bernstein, B. E. The NIH roadmap epigenomics mapping consortium. Nature Biotech. 28, 1045–1048 (2010).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Esteller, M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 16, R50–R59 (2007).
Esteller, M., Corn, P. G., Baylin, S. B. & Herman, J. G. A gene hypermethylation profile of human cancer. Cancer Res. 61, 3225–3229 (2001).
Javierre, B. M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Volkmar, M. et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 31, 1405–1426 (2012).
Fernandez, A. F. et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 22, 407–419 (2011).
Ahlquist, D. A. et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 142, 248–256; quiz e25–e26 (2012).
Kawakami, K. et al. Long interspersed nuclear element-1 hypomethylation is a potential biomarker for the prediction of response to oral fluoropyrimidines in microsatellite stable and CpG island methylator phenotype-negative colorectal cancer. Cancer Sci. 102, 166–174 (2011).
Cui, H. et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 62, 6442–6446 (2002).
Ito, Y. et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum. Mol. Genet. 17, 2633–2643 (2008).
Maxwell, A., McCudden, C. R., Wians, F. & Willis, M. S. Recent advances in the detection of prostate cancer using epigenetic markers in commonly collected laboratory samples. Lab. Med. 40, 171–178 (2009).
Feinberg, A. P. Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–440 (2007).
Berdasco, M. & Esteller, M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev. Cell 19, 698–711 (2010).
Lee, W. H. et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl Acad. Sci. USA 91, 11733–11737 (1994).
Yegnasubramanian, S. et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 64, 1975–1986 (2004).
Jerónimo, C. et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J. Natl Cancer Inst. 93, 1747–1752 (2001).
Bastian, P. J. et al. Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. Clin. Cancer Res. 11, 4097–4106 (2005).
Padar, A. et al. Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. Clin. Cancer Res. 9, 4730–4734 (2003).
Van Neste, L. et al. The epigenetic promise for prostate cancer diagnosis. Prostate 72, 1248–1261 (2011).
Nakayama, M. et al. Hypermethylation of the human glutathione S-transferase-π gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am. J. Pathol. 163, 923–933 (2003).
Goessl, C. et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 60, 5941–5945 (2000).
Reibenwein, J. et al. Promoter hypermethylation of GSTP1, AR, and 14-3-3σ in serum of prostate cancer patients and its clinical relevance. Prostate 67, 427–432 (2007).
Ellinger, J. et al. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate 68, 42–49 (2008).
Sunami, E. et al. Multimarker circulating DNA assay for assessing blood of prostate cancer patients. Clin. Chem. 55, 559–567 (2009).
Gonzalgo, M. L., Pavlovich, C. P., Lee, S. M. & Nelson, W. G. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin. Cancer Res. 9, 2673–2677 (2003).
Cairns, P. et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin. Cancer Res. 7, 2727–2730 (2001).
Goessl, C. et al. DNA-based detection of prostate cancer in urine after prostatic massage. Urology 58, 335–338 (2001).
Hoque, M. O. et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J. Clin. Oncol. 23, 6569–6575 (2005).
Rogers, C. G. et al. High concordance of gene methylation in post-digital rectal examination and post-biopsy urine samples for prostate cancer detection. J. Urol. 176, 2280–2284 (2006).
Wu, T. et al. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a meta-analysis. Br. J. Cancer 105, 65–73 (2011).
Ellinger, J. et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology 71, 161–167 (2008).
Lee, B. B. et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin. Cancer Res. 15, 6185–6191 (2009).
Lofton-Day, C. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 54, 414–423 (2008).
Warren, J. D. et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 9, 133 (2011).
Glockner, S. C. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 69, 4691–4699 (2009).
Balan´a, C. et al. Tumour and serum MGMT promoter methylation and protein expression in glioblastoma patients. Clin. Transl. Oncol. 13, 677–685 (2011).
Esteller, M. et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 59, 67–70 (1999).
Rosas, S. L. et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 61, 939–942 (2001).
Guerrero-Preston, R. et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. 4, 1061–1072 (2011).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nature Biotech. 28, 1057–1068 (2010).
Fraga, M. F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA 102, 10604–10609 (2005).
Rakyan, V. K. et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 7, e1002300 (2011).
Goate, A. & Hardy, J. Twenty years of Alzheimer's disease-causing mutations. J. Neurochem. 120 (Suppl. 1), 3–8 (2012).
Mastroeni, D., McKee, A., Grover, A., Rogers, J. & Coleman, P. D. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS ONE 4, e6617 (2009).
Heyn, H. et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl Acad. Sci. USA 109, 10522–10527 (2012).
Brock, M. V. DNA methylation markers and early recurrence in stage I lung cancer. N. Engl. J. Med. 358, 1118–1128 (2008).
Graff, J. R., Gabrielson, E., Fujii, H., Baylin, S. B. & Herman, J. G. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J. Biol. Chem. 275, 2727–2732 (2000).
Carmona, F. J. et al. Epigenetic disruption of cadherin-11 in human cancer metastasis. J. Pathol. 26 Jul 2012 (doi:10.1002/path.4011).
Lujambio, A. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA 105, 13556–13561 (2008).
Milani, L. et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood 115, 1214–1225 (2010).
Hinoue, T. et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 22, 271–281 (2011).
Zhuang, J. et al. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet. 8, e1002517 (2012).
Dedeurwaerder, S. et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 3, 726–741 (2011).
Fang, F. et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci. Transl. Med. 3, 75ra25 (2011).
Noushmehr, H. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Turcan, S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005).
Esteller, M., Hamilton, S. R., Burger, P. C., Baylin, S. B. & Herman, J. G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 59, 793–797 (1999).
Stupp, R. et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 28, 2712–2718 (2010).
Esteller, M. et al. Hypermethylation of the DNA repair gene O6-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J. Natl Cancer Inst. 94, 26–32 (2002).
Chakravarti, A. et al. Temozolomide-mediated radiation enhancement in glioblastoma: a report on underlying mechanisms. Clin. Cancer Res. 12, 4738–4746 (2006).
King, M.-C., Marks, J. H. & Mandell, J. B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003).
Esteller, M. et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl Cancer Inst. 92, 564–569 (2000).
Stefansson, O. A. et al. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 6, 638–649 (2011).
Silver, D. P. et al. Efficacy of neoadjuvant cisplatin in triple-negative breast Cancer J. Clin. Oncol. 28, 1145–1153 (2010).
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Veeck, J. et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J. Clin. Oncol. 28, e563–e564; author reply e565–e566 (2010).
Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA 89, 1827–1831 (1992).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008).
Lister, R. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Laurent, L. et al. Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331 (2010).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Li, Y. et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8, e1000533 (2010).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Bernstein, B. E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotech. 28, 1045–1048 (2010).
Bock, C. et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 (2011).
Irizarry, R. A. et al. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res. 18, 780–790 (2008).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genet. 41, 178–186 (2009).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010).
Kim, K. et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nature Biotech. 29, 1117–1119 (2011).
Doi, A. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature Genet. 41, 1350–1353 (2009).
McDonald, O. G., Wu, H., Timp, W., Doi, A. & Feinberg, A. P. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature Struct. Mol. Biol. 18, 867–874 (2011).
Serre, D., Lee, B. H. & Ting, A. H. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38, 391–399 (2010).
Taylor, B. S. et al. Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Disc. 1, 587–597 (2011).
Down, T. A. et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nature Biotech. 26, 779–785 (2008).
Jacinto, F. V., Ballestar, E. & Esteller, M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. BioTechniques 44, 35–43 (2008).
Feber, A. et al. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 21, 515–524 (2011).
Ball, M. P. et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature Biotech. 27, 361–368 (2009).
Maunakea, A. K. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Dedeurwaerder, S. et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics 3, 771–784 (2011).
Tost, J., Schatz, P., Schuster, M., Berlin, K. & Gut, I. G. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 31, e50 (2003).
Cocklin, R. R. & Wang, M. Identification of methylation and acetylation sites on mouse histone H3 using matrix-assisted laser desorption/ionization time-of-flight and nanoelectrospray ionization tandem mass spectrometry. J. Protein Chem. 22, 327–334 (2003).
Ehrich, M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA 102, 15785–15790 (2005).
Schatz, P., Dietrich, D. & Schuster, M. Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF. Nucleic Acids Res. 32, e167 (2004).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nature Biotech. 28, 1069–1078 (2010).
Yoo, C. B. et al. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 67, 6400–6408 (2007).
Kelly, W. K. et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J. Clin. Oncol. 23, 3923–3931 (2005).
Duvic, M. et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109, 31–39 (2007).
Garcia-Manero, G. et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood 111, 1060–1066 (2008).
Whittaker, S. J. et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 28, 4485–4491 (2010).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Bernt, K. M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011).
Krivtsov, A. V. et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14, 355–368 (2008).
Daigle, S. R. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Chekhun, V. F. et al. Role of DNA hypomethylation in the development of the resistance to doxorubicin in human MCF-7 breast adenocarcinoma cells. Cancer Lett. 231, 87–93 (2006).
Soengas, M. S. et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409, 207–211 (2001).
Iorns, E. et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell 13, 91–104 (2008).
Toyota, M. et al. Epigenetic inactivation of CHFR in human tumors. Proc. Natl Acad. Sci. USA 100, 7818–7823 (2003).
Fan, M. et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 66, 11954–11966 (2006).
Taniguchi, T. et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nature Med. 9, 568–574 (2003).
Dejeux, E. et al. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol. Cancer 9, 68 (2010).
Ibanez de Caceres, I. et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 29, 1681–1690 (2010).
Strathdee, G., MacKean, M. J., Illand, M. & Brown, R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene 18, 2335–2341 (1999).
Faller, W. J. et al. Metallothionein 1E is methylated in malignant melanoma and increases sensitivity to cisplatin-induced apoptosis. Melanoma Res. 20, 392–400 (2010).
Maier, S. et al. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients—technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur. J. Cancer 43, 1679–1686 (2007).
Syed, N. et al. Polo-like kinase Plk2 is an epigenetic determinant of chemosensitivity and clinical outcomes in ovarian cancer. Cancer Res. 71, 3317–3327 (2011).
Lee, J.-H. et al. Epigenetic alteration of PRKCDBP in colorectal cancers and its implication in tumor cell resistance to TNFα-induced apoptosis. Clin. Cancer Res. 17, 7551–7562 (2011).
Ramirez, J. L. et al. 14-3-3σ methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: the Spanish Lung Cancer Group. J. Clin. Oncol. 23, 9105–9112 (2005).
Ferreri, A. J. M. et al. Aberrant methylation in the promoter region of the reduced folate carrier gene is a potential mechanism of resistance to methotrexate in primary central nervous system lymphomas. Br. J. Haematol. 126, 657–664 (2004).
Tessema, M. et al. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene 12 Dec 2011 (doi:10.1038/onc.2011.577).
Ebert, M. P. A. et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N. Engl. J. Med. 366, 44–53 (2012).
Ai, L. et al. The transglutaminase 2 gene (TGM2), a potential molecular marker for chemotherapeutic drug sensitivity, is epigenetically silenced in breast cancer. Carcinogenesis 29, 510–518 (2008).
Shen, L. et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 67, 11335–11343 (2007).
Agrelo, R. et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc. Natl Acad. Sci. USA 103, 8822–8827 (2006).
Sabatino, M. A. et al. Down-regulation of the nucleotide excision repair gene XPG as a new mechanism of drug resistance in human and murine cancer cells. Mol. Cancer 9, 259 (2010).
Acknowledgements
The authors are supported by the European Research Council Advanced Grant EPINORC under the agreement no. 268626, the MICINN Project–SAF2011-22803, the European Community's Seventh Framework Programme (FP7/2007-2013) by the grant HEALTH-F5-2011-282510- BLUEPRINT, Fondo de Investigaciones Sanitarias Grant PI08-1345, the Dr. Josef Steiner Cancer Research Foundation Award, Botin Foundation, Cellex Foundation and the Health Department of the Catalan Government (Generalitat de Catalunya). M.E. is an Institucio Catalana de Recerca i Estudis Avançats (ICREA) Research Professor.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- CpG islands
-
CpG-rich regions of DNA that are often associated with the transcription start sites of genes and that are also found in gene bodies and intergenic regions.
- Personalized medicine
-
Therapeutic decisions based on genetic and epigenetic information of individual patients
- Prostate-specific antigen
-
(PSA). A serine protease of the kallikrein gene family that is secreted into seminal fluid by prostatic epithelial cells and is found in the serum. As it is almost exclusively a product of prostate cells, measurement in blood has proved to be useful as a tumour marker for diagnosis of prostate cancer and monitoring the effectiveness of treatment.
Rights and permissions
About this article
Cite this article
Heyn, H., Esteller, M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13, 679–692 (2012). https://doi.org/10.1038/nrg3270
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3270
This article is cited by
-
Comprehensive analysis of candidate signatures of long non-coding RNA LINC01116 and related protein-coding genes in patients with hepatocellular carcinoma
BMC Gastroenterology (2023)
-
A DNA methylation signature for the prediction of tumour recurrence in stage II colorectal cancer
British Journal of Cancer (2023)
-
Genome-wide methylation profiling identifies a novel gene signature for patients with synchronous colorectal cancer
British Journal of Cancer (2023)
-
P14.5 functions to inhibit cell migration and can be used as a prognostic marker in hepatocellular carcinoma
Genes & Genomics (2023)
-
A Self-attention Graph Convolutional Network for Precision Multi-tumor Early Diagnostics with DNA Methylation Data
Interdisciplinary Sciences: Computational Life Sciences (2023)