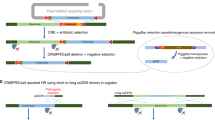
Abstract
Mouse models have become an invaluable tool for understanding human health and disease owing to our ability to manipulate the mouse genome exquisitely. Recent progress in genomic analysis has led to an increase in the number and type of disease-causing mutations detected and has also highlighted the importance of non-coding regions. As a result, there is increasing interest in creating 'genomically' humanized mouse models, in which entire human genomic loci are transferred into the mouse genome. The technical challenges towards achieving this aim are large but are starting to be tackled with success.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Nguyen, D. & Xu, T. The expanding role of mouse genetics for understanding human biology and disease. Dis. Model. Mech. 1, 56–66 (2008).
Fisher, E. M. C., Lana-Elola, E., Watson, S. D., Vassiliou, G. & Tybulewicz, V. L. J. New approaches for modelling sporadic genetic disease in the mouse. Dis. Model. Mech. 2, 446–453 (2009).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Giraldo, P. & Montoliu, L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 10, 83–103 (2001).
Heaney, J. D. & Bronson, S. K. Artificial chromosome-based transgenes in the study of genome function. Mamm. Genome 17, 791–807 (2006).
Li, L. P. et al. Transgenic mice with a diverse human T cell antigen receptor repertoire. Nature Med. 16, 1029–1034 (2010).
Gong, S. C., Kus, L. & Heintz, N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nature Protoc. 5, 1678–1696 (2010).
Johnson, S. J. & Wade-Martins, R. A BACwards glance at neurodegeneration: molecular insights into disease from LRRK2, SNCA and MAPT BAC-transgenic mice. Biochem. Soc. Trans. 39, 862–867 (2011).
Lonberg, N. et al. Antigen-specific human-antibodies from mice comprising 4 distinct genetic modifications. Nature 368, 856–859 (1994).
Heaney, J. D., Rettew, A. N. & Bronson, S. K. Tissue-specific expression of a BAC transgene targeted to the Hprt locus in mouse embryonic stem cells. Genomics 83, 1072–1082 (2004).
Prosser, H. M., Rzadzinska, A. K., Steel, K. P. & Bradley, A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol. Cell. Biol. 28, 1702–1712 (2008).
Luo, J. L. et al. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20, 320–328 (2001).
Song, H., Hollstein, M. & Xu, Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature Cell Biol. 9, 573–580 (2007).
Valenzuela, D. M. et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nature Biotech. 21, 652–659 (2003).
Rathinam, C. et al. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood 118, 3119–3128 (2011).
Rongvaux, A. et al. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc. Natl Acad. Sci. USA 108, 2378–2383 (2011).
Willinger, T. et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl Acad. Sci. USA 108, 2390–2395 (2011).
Murphy, A. VelocImmune: Immunoglobulin Variable Region Humanized Mice in Recombinant Antibodies for Immunotherapy (ed. Little, M.) 100–108 (Cambridge Univ. Press, UK, 2011).
Bouhassira, E. E., Westerman, K. & Leboulch, P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood 90, 3332–3344 (1997).
Khandelia, P., Yap, K. & Makeyev, E. V. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl Acad. Sci. USA 108, 12799–12804 (2011).
Liu, K. et al. Recombinase-mediated cassette exchange to rapidly and efficiently generate mice with human cardiac sodium channels. Genesis 44, 556–564 (2006).
Wallace, H. A. et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128, 197–209 (2007).
Hasegawa, M. et al. Quantitative prediction of human pregnane X receptor and cytochrome P450 3A4 mediated drug–drug interaction in a novel multiple humanized mouse line. Mol. Pharmacol. 80, 518–528 (2011).
Tybulewicz, V. L. & Fisher, E. M. New techniques to understand chromosome dosage: mouse models of aneuploidy. Hum. Mol. Genet. 15, R103–R109 (2006).
van der, W. L., Shaw-Smith, C. & Bradley, A. Chromosome engineering in ES cells. Methods Mol. Biol. 530, 49–77 (2009).
O'Doherty, A. et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309, 2033–2037 (2005).
Shinohara, T. et al. Mice containing a human chromosome 21 model behavioral impairment and cardiac anomalies of Down's syndrome. Hum. Mol. Genet. 10, 1163–1175 (2001).
Tomizuka, K. et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nature Genet. 16, 133–143 (1997).
Kazuki, Y. et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 18, 384–393 (2011).
Suzuki, N., Itou, T., Hasegawa, Y., Okazaki, T. & Ikeno, M. Cell to cell transfer of the chromatin-packaged human β-globin gene cluster. Nucleic Acids Res. 38, e33 (2010).
Kuroiwa, Y. et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nature Biotech. 18, 1086–1090 (2000).
Yamaguchi, S. et al. A method for producing transgenic cells using a multi-integrase system on a human artificial chromosome vector. PLoS ONE 6, e17267 (2011).
Nakano, M. et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 14, 507–522 (2008).
Kazuki, Y. & Oshimura, M. Human artificial chromosomes for gene delivery and the development of animal models. Mol. Ther. 19, 1591–1601 (2011).
Li, H. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011).
Gross, B. et al. Generation and characterization of a humanised PPARδ mouse model. Br. J. Pharmacol. 164, 192–208 (2011).
Uno, S. et al. CYP1A1 and CYP1A2 expression: comparing 'humanized' mouse lines and wild-type mice; comparing human and mouse hepatoma-derived cell lines. Toxicol. Appl. Pharmacol. 237, 119–126 (2009).
Wong, P. C., Cai, H. B., Borchelt, D. R. & Price, D. L. Genetically engineered mouse models of neurodegenerative diseases. Nature Neurosci. 5, 633–639 (2002).
Lehman, E. J. et al. Genetic background regulates β-amyloid precursor protein processing and β-amyloid deposition in the mouse. Hum. Mol. Genet. 12, 2949–2956 (2003).
Anastassiadis, K. et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model. Mech. 2, 508–515 (2009).
Tomizuka, K. et al. Double trans-chromosomic mice: maintenance of two individual human chromosome fragments containing Ig heavy and κ loci and expression of fully human antibodies. Proc. Natl Acad. Sci. USA 97, 722–727 (2000).
Animals containing human material. The Academy of Medical Sciences [online], (2011).
Vernimmen, D. et al. Polycomb eviction as a new distant enhancer function. Genes Dev. 25, 1583–1588 (2011).
Vernimmen, D. et al. Chromosome looping at the human α-globin locus is mediated via the major upstream regulatory element (HS-40). Blood 114, 4253–4260 (2009).
Wilson, M. D. et al. Species-specific transcription in mice carrying human chromosome 21. Science 322, 434–438 (2008).
Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Hawkins, P. N., Myers, M. J., Epenetos, A. A., Caspi, D. & Pepys, M. B. Specific localization and imaging of amyloid deposits in vivo using 123I-labeled serum amyloid P component. J. Exp. Med. 167, 903–913 (1988).
Grad, L. I. et al. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc. Natl Acad. Sci. USA 108, 16398–16403 (2011).
Dorner, M. et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 474, 208–211 (2011).
Enard, W. et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971 (2009).
Stahl, P. D. & Wainszelbaum, M. J. Human-specific genes may offer a unique window into human cell signaling. Sci. Signal. 2, e59 (2009).
Schorderet, P. & Duboule, D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet. 7, e1002071 (2011).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell 129, 1311–1323 (2007).
Yeretssian, G. et al. Gender differences in expression of the human caspase-12 long variant determines susceptibility to Listeria monocytogenes infection. Proc. Natl Acad. Sci. USA 106, 9016–9020 (2009).
Acknowledgements
A.D., R.B.-S., V.J.L.T., A.J.H.S. and E.M.C.F. were funded by the UK Medical Research Council, and V.L.J.T. and E.M.C.F. also received funding from the Wellcome Trust and the AnEUploidy consortium, a European Union FP6 Integrated Project. We thank M. Pepys, G. Taylor and S. Wood for information about serum amyloid P (SAP) and J.-F. Bureau, P. D'Eustachio, D. Hughes, R. Kumar, M. Landrum, J. Milner and H. Noyes for information about human-specific genes. We apologize if important papers have been omitted owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Conditional mutations
-
Mutated sequences that are expressed under experimental control. For example, in a conditional knockout, deletion of a crucial exon that is flanked by recombination sites can be induced by expressing a recombinase that is under the control of a tissue-specific promoter.
- Gene targeting
-
A method of exchanging genetic information from a donor vector into a recipient genome by exploiting a DNA repair mechanism that recognizes homology between the donor DNA and the recipient locus. It results in the replacement of the original genomic sequence with the donor sequence.
- Genome-wide association studies
-
(GWASs). Whole-genome scans that are designed to test the statistical association between finely spaced polymorphic genetic markers and traits under study. Large numbers of phenotypically well-classified DNA samples are required to have sufficient statistical power to detect such associations.
- Inducible mutations
-
A class of conditional mutation that is expressed by using a ligand-inducible recombinase. Inducible mutations allow recombinase-driven changes to be induced at a particular time point by delivering the ligand into the animal by injection, diet or using viral vectors.
- Knock-in mice
-
Mice carrying a targeted replacement or insertion. This could be generated by an exchange of nucleotide sequence to change the encoded protein sequence or by an insertion to create a tagged protein.
- Knockout mice
-
Mice in which the functional protein-coding capacity of a particular gene has been disrupted. This could be generated by a targeted deletion, gene trap, or targeted conditional knockout.
- Knockout Mouse Project
-
(KOMP). A programme funded by the US National Institutes of Health (NIH) contributing to the International Mouse Knockout Consortium (IKMC) to generate a null mutation in every mouse gene. Other contributors are the Texas Institute for Genomic Medicine (TIGM), the North American Conditional Mouse Mutagenesis Program (NorCOMM) and the European Conditional Mouse Mutagenesis Programme (EUCOMM).
- Microcell-mediated chromosome transfer
-
(MMCT). A method for transferring entire chromosomes or chromosome fragments from a donor cell line into a recipient cell line.
- Minigene
-
A gene construct that contains fewer exons and introns than its original full gene counterpart. Minigenes have been used to investigate regulatory sequences but may also be used because of their more convenient size.
- Pronuclear injection
-
A process by which DNA is added to the genome, generally to create transgenic mice. Linearized DNA is injected into one of the two pronuclei of the fertilized egg, where it is stably incorporated into the genome. The eggs are then developed to term in a pseudopregnant female mouse.
- Recombineering
-
Derived from 'recombination-mediated genetic engineering', this process uses recombination functions encoded by λ-phages to mediate in vivo recombination using short stretches of homologous sequence (30–50 nucleotides long) that can be readily appended to the termini of any desired DNA fragment.
- Transchromosomic mouse strains
-
A mouse strain into which either an entire chromosome or a chromosome fragment from another species has been transferred. This chromosome or chromosome fragment may be freely segregating or inserted into an existing mouse chromosome.
- Xenogeneic transplantation
-
The transplantation of tissues or cells derived from one species into another. For example, the introduction of human stem cells into an immune-deficient mouse strain.
Rights and permissions
About this article
Cite this article
Devoy, A., Bunton-Stasyshyn, R., Tybulewicz, V. et al. Genomically humanized mice: technologies and promises. Nat Rev Genet 13, 14–20 (2012). https://doi.org/10.1038/nrg3116
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3116
This article is cited by
-
Genetic advancements in obesity management and CRISPR–Cas9-based gene editing system
Molecular and Cellular Biochemistry (2023)
-
Humanized Mice for Infectious and Neurodegenerative disorders
Retrovirology (2021)
-
Dissection and function of autoimmunity-associated TNFAIP3 (A20) gene enhancers in humanized mouse models
Nature Communications (2018)
-
Moving toward a higher efficiency of microcell-mediated chromosome transfer
Molecular Therapy - Methods & Clinical Development (2016)
-
Retargeting of microcell fusion towards recipient cell-oriented transfer of human artificial chromosome
BMC Biotechnology (2015)