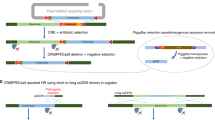
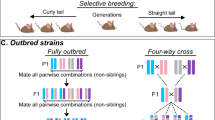
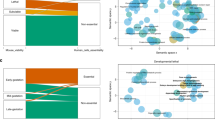
Key Points
-
The mouse is the leading mammalian model organism for basic genetic research and to study human diseases. The current mouse mutant resources already contain mutations for essentially all genes and markers, but many of these mutations still need to be established in mouse lines as a free resource for the scientific community.
-
To maximize the potential of the mouse as a model organism, we now face challenges that go beyond simply establishing a mutant line for every gene. The focus of genetic research for the next generation of mouse models can be considered at three main levels: the genotype, the phenotype and what we term the envirotype.
-
At the genotype level, mutations are required that are more closely related to human mutations and genetic variants.
-
At the phenotype level, systematic (each mutant mouse line) and systemic (examining all organ systems) analysis of mutant mouse lines is required.
-
Several mouse clinics have provided essential phenotype data that is fundamental for the analysis of primary and secondary effects of mutations. The capacities of these mouse clinics will need to be increased to make significant progress at a faster pace.
-
The analysis of envirotypes (which are sets of exogenous factors that affect the phenotype) will be essential for accurately modelling human diseases in the mouse, but this research is just beginning.
-
A map of exogenous factors and their effects on the mammalian genome will be required. The extent to which the effects of particular envirotypes are the same in mice and humans remains to be determined.
Abstract
The mouse is the leading mammalian model organism for basic genetic research and for studying human diseases. Coordinated international projects are currently in progress to generate a comprehensive map of mouse gene functions — the first for any mammalian genome. There are still many challenges ahead to maximize the value of the mouse as a model, particularly for human disease. These involve generating mice that are better models of human diseases at the genotypic level, systemic (assessing all organ systems) and systematic (analysing all mouse lines) phenotyping of existing and new mouse mutant resources, and assessing the effects of the environment on phenotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nadeau, J. H. et al. Sequence interpretation. Functional annotation of mouse genome sequences. Science 291, 1251–1255 (2001).
Austin, C. P. et al. The knockout mouse project. Nature Genet. 36, 921–924 (2004).
Auwerx, J. et al. The European dimension for the mouse genome mutagenesis program. Nature Genet. 36, 925–927 (2004).
Patten, B. C. in Eco Targets, Goal Functions, and Orientors (eds Muller, F. & Leupelt, M.) 137–160 (Springer, Berlin, 1998). We believe that this publication introduced the term 'envirotype'. In particular, it is mentioned that “the genotype–phenotype pair of classical genetics is an incomplete specification of determinate reproduction; an external envirotype is needed to complete the mechanism.”
Bult, C. J., Eppig, J. T., Kadin, J. A., Richardson, J. E. & Blake, J. A. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36, D724–D728 (2008).
Paigen, K. One hundred years of mouse genetics: an intellectual history. II. The molecular revolution (1981–2002). Genetics 163, 1227–1235 (2003). References 6 and 7 give an excellent historical overview of 100 years of mouse genetics, from its beginning to the genomic era.
Paigen, K. One hundred years of mouse genetics: an intellectual history. I. The classical period (1902–1980). Genetics 163, 1–7 (2003).
Qiu, J. Animal research: mighty mouse. Nature 444, 814–816 (2006).
Collins, F. S., Finnell, R. H., Rossant, J. & Wurst, W. A new partner for the international knockout mouse consortium. Cell 129, 235 (2007).
Collins, F. S., Rossant, J. & Wurst, W. A mouse for all reasons. Cell 128, 9–13 (2007).
Hansen, G. M. et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 18, 1670–1679 (2008).
Gondo, Y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nature Rev. Genet. 9, 803–810 (2008).
Carlson, C. M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Keng, V. W. et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nature Biotechnol. 27, 264–274 (2009).
Rosenthal, N. & Brown, S. The mouse ascending: perspectives for human-disease models. Nature Cell Biol. 9, 993–999 (2007). A comprehensive and insightful recent review of the genetic tools available for the mouse and the challenges that mouse models of human diseases are facing.
Gieger, C. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4, e1000282 (2008).
McCarroll, S. A. & Altshuler, D. M. Copy-number variation and association studies of human disease. Nature Genet. 39, S37–S42 (2007).
McCarroll, S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genet. 40, 1166–1174 (2008).
Justice, M. J., Noveroske, J. K., Weber, J. S., Zheng, B. & Bradley, A. Mouse ENU mutagenesis. Hum. Mol. Genet. 8, 1955–1963 (1999).
Soewarto, D., Klaften, M. & Rubio-Aliaga, I. Features and strategies of ENU mouse mutagenesis. Curr. Pharm. Biotechnol. 10, 198–213 (2009).
Nolan, P. M. et al. Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm. Genome 11, 500–506 (2000).
Nolan, P. M. et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nature Genet. 25, 440–443 (2000).
Hrabe de Angelis, M. H. et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nature Genet. 25, 444–447 (2000).
Balling, R. ENU mutagenesis: analyzing gene function in mice. Annu. Rev. Genomics Hum. Genet. 2, 463–492 (2001).
Vreugde, S. et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nature Genet. 30, 257–258 (2002). This paper of an ENU-induced mouse model was published along with an article that describes patients with the same progressive deafness phenotype caused by a mutation in the homologous human gene.
Lisse, T. S. et al. ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet. 4, e7 (2008).
Klaften, M. & Hrabe de Angelis, M. ARTS: a web-based tool for the set-up of high-throughput genome-wide mapping panels for the SNP genotyping of mouse mutants. Nucleic Acids Res. 33, W496–W500 (2005).
Augustin, M. et al. Efficient and fast targeted production of murine models based on ENU mutagenesis. Mamm. Genome 16, 405–413 (2005).
Coghill, E. L. et al. A gene-driven approach to the identification of ENU mutants in the mouse. Nature Genet. 30, 255–256 (2002).
Michaud, E. J. et al. Efficient gene-driven germ-line point mutagenesis of C57BL/6J mice. BMC Genomics 6, 164 (2005).
Quwailid, M. M. et al. A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm. Genome 15, 585–591 (2004).
Sakuraba, Y. et al. Molecular characterization of ENU mouse mutagenesis and archives. Biochem. Biophys. Res. Commun. 336, 609–616 (2005).
Sakuraba, Y. et al. Identification and characterization of new long conserved noncoding sequences in vertebrates. Mamm. Genome 19, 703–712 (2008).
Takahasi, K. R., Sakuraba, Y. & Gondo, Y. Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Mol. Biol. 8, 52 (2007).
Herault, Y., Rassoulzadegan, M., Cuzin, F. & Duboule, D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nature Genet. 20, 381–384 (1998).
Olson, L. E., Richtsmeier, J. T., Leszl, J. & Reeves, R. H. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science 306, 687–690 (2004).
Kmita, M., Fraudeau, N., Herault, Y. & Duboule, D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420, 145–150 (2002).
Couzin, J. RNA interference. Mini RNA molecules shield mouse liver from hepatitis. Science 299, 995 (2003).
Kunath, T. Transgenic RNA interference to investigate gene function in the mouse. Methods Mol. Biol. 461, 165–186 (2008).
Raoul, C. et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Med. 11, 423–428 (2005).
Bibikova, M., Golic, M., Golic, K. G. & Carroll, D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169–1175 (2002).
Lloyd, A., Plaisier, C. L., Carroll, D. & Drews, G. N. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl Acad. Sci. USA 102, 2232–2237 (2005).
Zeevi, V., Tovkach, A. & Tzfira, T. Increasing cloning possibilities using artificial zinc finger nucleases. Proc. Natl Acad. Sci. USA 105, 12785–12790 (2008).
Mani, M., Kandavelou, K., Dy, F. J., Durai, S. & Chandrasegaran, S. Design, engineering, and characterization of zinc finger nucleases. Biochem. Biophys. Res. Commun. 335, 447–457 (2005).
Steuber-Buchberger, P., Wurst, W. & Kuhn, R. Simultaneous Cre-mediated conditional knockdown of two genes in mice. Genesis 46, 144–151 (2008).
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008).
Hitz, C., Steuber-Buchberger, P., Delic, S., Wurst, W. & Kuhn, R. Generation of shRNA transgenic mice. Methods Mol. Biol. 530, 1–29 (2009).
Echeverri, C. J. et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nature Methods 3, 777–779 (2006).
Meng, X., Noyes, M. B., Zhu, L. J., Lawson, N. D. & Wolfe, S. A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnol. 26, 695–701 (2008).
Yan, Z., Sun, X. & Engelhardt, J. F. Progress and prospects: techniques for site-directed mutagenesis in animal models. Gene Ther. 19 Feb 2009 (doi:10.1038/gt.2009.16).
Matera, I. et al. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum. Mol. Genet. 17, 2118–2131 (2008).
Mohan, S., Baylink, D. J. & Srivastava, A. K. A chemical mutagenesis screen to identify modifier genes that interact with growth hormone and TGF-beta signaling pathways. Bone 42, 388–395 (2008).
Rubio-Aliaga, I. et al. A genetic screen for modifiers of the delta1-dependent Notch signaling function in the mouse. Genetics 175, 1451–1463 (2007).
Dietrich, W. F. et al. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75, 631–639 (1993). A hallmark article on the genetic mapping of the first quantitative trait gene or modifier affecting a mouse model for human disease.
Erickson, R. P. Mouse models of human genetic disease: which mouse is more like a man? Bioessays 18, 993–998 (1996).
Gregorova, S. et al. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 18, 509–515 (2008).
Rogner, U. C. & Avner, P. Congenic mice: cutting tools for complex immune disorders. Nature Rev. Immunol. 3, 243–252 (2003).
Matin, A., Collin, G. B., Asada, Y., Varnum, D. & Nadeau, J. H. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nature Genet. 23, 237–240 (1999).
Nadeau, J. H., Singer, J. B., Matin, A. & Lander, E. S. Analysing complex genetic traits with chromosome substitution strains. Nature Genet. 24, 221–225 (2000).
Singer, J. B. et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304, 445–448 (2004).
Singer, J. B., Hill, A. E., Nadeau, J. H. & Lander, E. S. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics 169, 855–862 (2005).
Grattan, M., Mi, Q. S., Meagher, C. & Delovitch, T. L. Congenic mapping of the diabetogenic locus Idd4 to a 5.2-cM region of chromosome 11 in NOD mice: identification of two potential candidate subloci. Diabetes 51, 215–223 (2002).
Hill, N. J. et al. NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes 49, 1744–1747 (2000).
Lamhamedi-Cherradi, S. E. et al. Further mapping of the Idd5.1 locus for autoimmune diabetes in NOD mice. Diabetes 50, 2874–2878 (2001).
Shao, H. et al. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl Acad. Sci. USA 105, 19910–19914 (2008). An insightful article on the frequent occurrence of QTLs in the rodent genome and how the interaction of QTLs is neither simple nor additive.
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Churchill, G. A. et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature Genet. 36, 1133–1137 (2004).
Iraqi, F. A., Churchill, G. & Mott, R. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm. Genome 19, 379–381 (2008).
Chesler, E. J. et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19, 382–389 (2008).
Davisson, M. FIMRe: Federation of International Mouse Resources: global networking of resource centers. Mamm. Genome 17, 363–364 (2006).
Davis, M. M. A Prescription for human immunology. Immunity 29, 835–838 (2008).
von Herrath, M. G. & Nepom, G. T. Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J. Exp. Med. 202, 1159–1162 (2005).
Nishie, W. et al. Humanization of autoantigen. Nature Med. 13, 378–383 (2007).
Traggiai, E. et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science 304, 104–107 (2004).
Cocco, M. et al. CD34+ cord blood cell-transplanted Rag2−/−γc−/− mice as a model for Epstein–Barr virus infection. Am. J. Pathol. 173, 1369–1378 (2008).
Gorantla, S. et al. Human immunodeficiency virus type 1 pathobiology studied in humanized BALB/c-Rag2−/−γc−/− mice. J. Virol. 81, 2700–2712 (2007).
Proia, D. A. & Kuperwasser, C. Reconstruction of human mammary tissues in a mouse model. Nature Protoc. 1, 206–214 (2006).
Gailus-Durner, V. et al. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nature Methods 2, 403–404 (2005).
Brown, S. D., Chambon, P. & de Angelis, M. H. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nature Genet. 37, 1155 (2005).
Green, E. C. et al. EMPReSS: European mouse phenotyping resource for standardized screens. Bioinformatics 21, 2930–2931 (2005).
Mallon, A. M., Blake, A. & Hancock, J. M. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 36, D715–D718 (2008).
Woychik, R. P., Klebig, M. L., Justice, M. J., Magnuson, T. R. & Avner, E. D. Functional genomics in the post-genome era. Mutat. Res. 400, 3–14 (1998).
Horsch, M. et al. Systematic gene expression profiling of mouse model series reveals coexpressed genes. Proteomics 8, 1248–1256 (2008).
Frey, I. M. et al. Profiling at mRNA, protein, and metabolite levels reveals alterations in renal amino acid handling and glutathione metabolism in kidney tissue of Pept2−/− mice. Physiol. Genomics 28, 301–310 (2007).
Ntziachristos, V., Culver, J. P. & Rice, B. W. Small-animal optical imaging. J. Biomed. Opt. 13, 011001 (2008).
Niedre, M. J. et al. Early photon tomography allows fluorescence detection of lung carcinomas and disease progression in mice in vivo. Proc. Natl Acad. Sci. USA 105, 19126–19131 (2008).
Ahting, U. et al. Neurological phenotype and reduced lifespan in heterozygous Tim23 knockout mice, the first mouse model of defective mitochondrial import. Biochim. Biophys. Acta 9 Dec 2008 (doi:10.1016/j.bbabio.2008.12.001).
Soker, T. et al. Pleiotropic effects in Eya3 knockout mice. BMC Dev. Biol. 8, 118 (2008).
Hoelter, S. M. et al. “Sighted C3H” mice — a tool for analysing the influence of vision on mouse behaviour? Front. Biosci. 13, 5810–5823 (2008).
Schmidt, S. et al. Deletion of glucose transporter GLUT8 in mice increases locomotor activity. Behav. Genet. 38, 396–406 (2008).
Fuchs, H. et al. Phenotypic characterization of mouse models for bone-related diseases in the German Mouse Clinic. J. Musculoskelet. Neuronal Interact. 8, 13–14 (2008).
Bender, A. et al. Creatine improves health and survival of mice. Neurobiol. Aging 29, 1404–1411 (2008).
Vauti, F. et al. The mouse Trm1-like gene is expressed in neural tissues and plays a role in motor coordination and exploratory behaviour. Gene 389, 174–185 (2007).
Barrantes Idel, B. et al. Generation and characterization of dickkopf3 mutant mice. Mol. Cell Biol. 26, 2317–2326 (2006).
Colucci-Guyon, E., Gimenez, Y. R. M., Maurice, T., Babinet, C. & Privat, A. Cerebellar defect and impaired motor coordination in mice lacking vimentin. Glia 25, 33–43 (1999).
Colucci-Guyon, E. et al. Mice lacking vimentin develop and reproduce without an obvious phenotype. Cell 79, 679–694 (1994).
Eckes, B. et al. Impaired wound healing in embryonic and adult mice lacking vimentin. J. Cell Sci. 113, 2455–2462 (2000).
Henrion, D. et al. Impaired flow-induced dilation in mesenteric resistance arteries from mice lacking vimentin. J. Clin. Invest. 100, 2909–2914 (1997).
Schiffers, P. M. et al. Altered flow-induced arterial remodeling in vimentin-deficient mice. Arterioscler. Thromb. Vasc. Biol. 20, 611–616 (2000).
Terzi, F. et al. Reduction of renal mass is lethal in mice lacking vimentin. Role of endothelin-nitric oxide imbalance. J. Clin. Invest. 100, 1520–1528 (1997).
Welsh, E., Jirotka, M. & Gavaghan, D. Post-genomic science: cross-disciplinary and large-scale collaborative research and its organizational and technological challenges for the scientific research process. Philos. Transact. A Math. Phys. Eng. Sci. 364, 1533–1549 (2006). A sociological study on the far-reaching impact that the advent of 'big science' in life science research is beginning to have, for example, in the areas of organizational cultures, working practice, rewarding systems, education and communication technology.
Paigen, K. & Eppig, J. T. A mouse phenome project. Mamm. Genome 11, 715–717 (2000).
Bogue, M. Mouse Phenome Project: understanding human biology through mouse genetics and genomics. J. Appl. Physiol. 95, 1335–1337 (2003).
Bogue, M. A. & Grubb, S. C. The Mouse Phenome Project. Genetica 122, 71–74 (2004).
Bogue, M. A., Grubb, S. C., Maddatu, T. P. & Bult, C. J. Mouse Phenome Database (MPD). Nucleic Acids Res. 35, D643–D649 (2007).
Hancock, J. M. & Mouse Phenotype Database Integration Consortium. Integration of mouse phenome data resources. Mamm. Genome 18, 157–163 (2007).
Brown, S. D., Hancock, J. M. & Gates, H. Understanding mammalian genetic systems: the challenge of phenotyping in the mouse. PLoS Genet. 2, e118 (2006).
Richter, S. H., Garner, J. P. & Wurbel, H. Environmental standardization: cure or cause of poor reproducibility in animal experiments? Nature Methods 6, 257–261 (2009).
Valérie Gailus-Durner et al. in Gene Knockout Protocols 2nd edn Vol. 530 (eds Wurst, W. & Kühn, R.) 436–509 (Humana Press, New Jersey, 2009).
Acknowledgements
The authors are funded through the German Ministry of Science and Education and the European Commission (grant numbers: 01GS0850, LSHG-2006-037188, 211414, LSHG-CT-2006-518240, MRTN-CT-2006-035468).
Author information
Authors and Affiliations
Corresponding authors
Related links
Related links
FURTHER INFORMATION
Australian Phenomics Facility (APF)
Charles River's phenotyping screens, Massachusetts
Comparative Pathology Laboratory (CPL)
Institut Clinique de la Souris (ICS)
Jackson Laboratory Phenotyping Services
Laboratory Animal Sciences Program (LASP)
Mammalian Genetics Phenotyping
Mouse Genetics Programme — Phenotyping
Mouse Phenotyping Shared Resource (MPSR)
Research Animal Diagnostic Laboratory (RADIL)
Toronto Centre for Phenogenomics (TCP)
Glossary
- Genotype
-
A description of the endogenous genetic information carried by an organism, as distinguished from its physical appearance (its phenotype) and external environmental factors (its envirotype).>
- Phenotype
-
A description of any observable (macroscopic, microscopic or molecular) trait of an individual with respect to some inherited characteristic.
- Envirotype
-
A description of factors that are exogenous to the organism. The environmental code and the genetic code together affect the phenotype.
- Pleiotropic
-
A situation in which a single gene has an effect on two or more distinct phenotypic characters.
- Transposon
-
A type of mobile genetic element that consists of DNA that can move to new genomic locations conservatively (without replicating itself) or replicatively (by moving a copy of itself).
- QTL
-
Genetic locus or chromosomal region that contributes to the variability in complex quantitative traits (such as body weight), as identified by statistical analysis. Quantitative traits are typically affected by several genes and by the environment.
- Genotype-driven mutagenesis
-
A reverse genetic approach that starts with the targeted mutagenesis of a known gene or marker sequence. The gene targeting is followed by the analysis of the mutant phenotype. This approach is generally based on a hypothesis about a potential function of the mutated gene.
- Phenotype-driven mutagenesis
-
A forward genetic approach that starts with the identification of a mutant phenotype caused by a random mutation in the genome. The identification of the mutated gene or marker is subsequent to the identification of the mutant mouse line. This approach makes no assumption of which genes may underlie a disease.
- SNP
-
A type of polymorphism in which genomic segments differ by a single base pair.
- Copy number variant
-
A type of polymorphism in which a segment of genomic DNA is present at a different copy number with respect to a reference genome.
- N-ethyl-N-nitrosourea
-
A chemical mutagen that introduces point mutations in spermatogonia of male mice with high efficiency. It can be used as a mutagen in gene-driven and phenotype-driven mutagenesis.
- Zinc-finger nuclease
-
Synthetic protein composed of a nonspecific DNA-cleaving domain and a highly specific DNA-binding domain, which comprises a string of zinc-finger motifs. Zinc-finger nucleases and subsequent DNA repair by homologous recombination can be used to mutagenize genes.
- Off-target effect
-
These effects may compromise the specificity of RNAi and can occur if there is sequence identity between the small interfering RNA and random mRNA transcripts, causing knockdown of the expression of non-targeted genes.
- Redundancy
-
When two genes can fulfil an equivalent function. Because gene functions are frequently pleiotropic, redundancy is often partial, with two genes having overlapping rather than equivalent functions.
- Epistasis
-
The interaction between different genes that affect the same trait. Epistasis takes place when the phenotype of one genetic allele (mutant or natural variant) is modified by one or several other genes (also called modifier genes), such that the joint phenotype differs from the one that would be produced if the two genes were acting independently.
- Sensitized mutagenesis screen
-
A phenotype-driven mutagenesis screen in which mice carrying a targeted mutation are bred with N-ethyl-N-nitrosourea-treated males in order to provide a sensitized system for detecting dominant modifier mutations.
- Consomic
-
Describes a mouse strain that is produced by a breeding strategy in which recombinants between two inbred strains are backcrossed to produce a strain that carries a single chromosome from one strain on the genetic background of the other.
- Congenic
-
Describes a mouse strain that is produced by a breeding strategy in which recombinants between two inbred strains are backcrossed to produce a strain that carries a single genomic segment from one strain on the genetic background of the other.
Rights and permissions
About this article
Cite this article
Beckers, J., Wurst, W. & de Angelis, M. Towards better mouse models: enhanced genotypes, systemic phenotyping and envirotype modelling. Nat Rev Genet 10, 371–380 (2009). https://doi.org/10.1038/nrg2578
Issue Date:
DOI: https://doi.org/10.1038/nrg2578
This article is cited by
-
Artificial Intelligence for Risk Assessment on Primary Prevention of Coronary Artery Disease
Current Cardiovascular Risk Reports (2023)
-
Analyzing Multiple Phenotypes Based on Principal Component Analysis
Acta Mathematicae Applicatae Sinica, English Series (2022)
-
Enviromics in breeding: applications and perspectives on envirotypic-assisted selection
Theoretical and Applied Genetics (2021)
-
Digital envirotyping: quantifying environmental determinants of health and behavior
npj Digital Medicine (2020)
-
Reply to ‘It is time for an empirically informed paradigm shift in animal research’
Nature Reviews Neuroscience (2020)