Key Points
-
The left–right asymmetrical placement of internal organs that characterizes the vertebrate body plan is established during embryogenesis by complex genetic and epigenetic cascades.
-
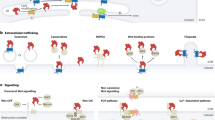
In all vertebrates that have been analysed so far, the expression of transcripts that encode the transforming growth factor-β (TGFB)-like signal NODAL is restricted to the left lateral plate mesoderm and this correlates with the establishment of proper left–right organ asymmetries.
-
The embryo node (the organizer in Xenopus laevis, Hensen's node in birds and mammals) or its derivatives (Kupffer's vesicle in teleost fish) have key roles as organizing centres for the determination of left–right visceral asymmetries.
-
In the mouse, the rotation of monocilia that project from the ventral side of the node creates a leftward flow of extracellular fluid that is necessary for proper left–right patterning (the nodal flow). Recent experimental and mathematical studies have shown that this directional flow can be generated de novo by cilia rotation in the absence of pre-existing laterality cues. Therefore, the nodal flow could be the initial symmetry-breaking event in the mouse embryo.
-
Recent reports also show that the nodal flow takes place in embryos other than the mouse, including those of zebrafish, medaka fish and the rabbit. At least in zebrafish, the nodal flow is required for normal left–right asymmetrical patterning.
-
We discuss the possibility that the nodal flow might amplify pre-existing laterality cues, rather than generate left–right asymmetry de novo, because further requirements for left–right patterning have been identified (at least in the zebrafish) that predate the establishment of the nodal flow.
-
With the exception of the mouse, early requirements for correct left–right patterning have been identified in all the other main vertebrate model organisms (X. laevis, chicks and zebrafish), including gap junction communications, differences in H+/K+-ATPase activity, asymmetrical gene expression, and Notch signalling. Data from various model organisms might provide insights into how these mechanisms relate to each other.
-
Recent evidence shows that the mechanisms that control left–right asymmetries in inner organ placement and those that generate the bilaterally symmetrical outer body wall are integrated at the level of the node.
Abstract
Although vertebrates seem to be essentially bilaterally symmetrical on the exterior, there are numerous interior left–right asymmetries in the disposition and placement of internal organs. These asymmetries are established during embryogenesis by complex epigenetic and genetic cascades. Recent studies in a range of model organisms have made important progress in understanding how this laterality information is generated and conveyed to large regions of the embryo. Both commonalities and divergences are emerging in the mechanisms that different vertebrates use in left–right axis specification. Recent evidence also provides intriguing links between the establishment of left–right asymmetries and the symmetrical elongation of the anterior–posterior axis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Palmer, A. R. Symmetry breaking and the evolution of development. Science 306, 828–833 (2004).
Capdevila, J., Vogan, K. J., Tabin, C. J. & Izpisúa Belmonte, J. C. Mechanisms of left–right determination in vertebrates. Cell 101, 9–21 (2000).
Mercola, M. & Levin, M. Left–right asymmetry determination in vertebrates. Annu. Rev. Cell Dev. Biol. 17, 779–805 (2001).
Hamada, H., Meno, C., Watanabe, D. & Saijoh, Y. Establishment of vertebrate left–right asymmetry. Nature Rev. Genet. 3, 103–113 (2002).
Bisgrove, B. W., Morelli, S. H. & Yost, H. J. Genetics of human laterality disorders: Insights from vertebrate model systems. Annu. Rev. Genomics Hum. Genet. 4, 1–32 (2003).
Levin, M. Left–right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 122, 3–25 (2005).
Brown, N. A. & Wolpert, L. The development of handedness in left/right asymmetry. Development 109, 1–9 (1990).
Tabin, C. J. & Vogan, K. J. A two-cilia model for vertebrate left–right axis specification. Genes Dev. 17, 1–6 (2003).
McGrath, J. & Brueckner, M. Cilia are at the heart of vertebrate left–right asymmetry. Curr. Opin. Genet. Dev. 13, 385–392 (2003).
Mercola, M. Left–right asymmetry: nodal points. J. Cell Sci. 116, 3251–3257 (2003).
Yost, H. J. Left–right asymmetry: nodal cilia make and catch a wave. Curr. Biol. 13, R808–R809 (2003).
Cooke, J. The evolutionary origins and significance of vertebrate left–right organisation. Bioessays 26, 413–421 (2004).
Wood, W. B. The left–right polarity puzzle: determining embryonic handedness. PLoS Biol. 3, e292 (2005).
Essner, J. J. et al. Conserved function for embryonic nodal cilia. Nature 418, 37–38 (2002).
Levin, M. Motor protein control of ion flux is an early step in embryonic left–right asymmetry. Bioessays 25, 1002–1010 (2003).
Levin, M., Johnson, R. L., Stern, C. D., Kuehn, M. & Tabin, C. A molecular pathway determining left–right asymmetry in chick embryogenesis. Cell 82, 803–814 (1995). This is the first report of genes that are expressed LR asymmetrically. NODAL and SHH are among the genes identified in this paper as having a role in LR patterning in the chick.
Raya, A. & Izpisúa Belmonte, J. C. Unveiling the establishment of left–right asymmetry in the chick embryo. Mech. Dev. 121, 1043–1054 (2004).
Hamburger, V. & Hamilton, H. L. A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951).
Levin, M., Thorlin, T., Robinson, K., Nogi, T. & Mercola, M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell 111, 77–89 (2002). The authors identify an LR difference in H+/K+-ATPase activity as an early epigenetic requirement for LR patterning of chick and X. laevis embryos.
Kawakami, Y., Raya, A., Raya, R. M., Rodriguez Esteban, C. & Izpisua Belmonte, J. C. Retinoic acid signaling links left–right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature 435, 165–171 (2005). This study characterized the early steps of LR specification in the zebrafish. Together with references 85 and 86, this paper uncovers a link between the pathways that control LR asymmetrical patterning and bilaterally symmetrical somitogenesis.
Duboc, V., Rottinger, E., Lapraz, F., Besnardeau, L. & Lepage, T. Left–right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev. Cell 9, 147–158 (2005).
Levin, M. & Nascone, N. Two molecular models of initial left–right asymmetry generation. Med. Hypotheses 49, 429–435 (1997).
Fukumoto, T., Kema, I. P. & Levin, M. Serotonin signaling is a very early step in patterning of the left–right axis in chick and frog embryos. Curr. Biol. 15, 794–803 (2005).
Sarmah, B., Latimer, A. J., Appel, B. & Wente, S. R. Inositol polyphosphates regulate zebrafish left–right asymmetry. Dev. Cell 9, 133–145 (2005).
Raya, A. et al. Notch activity acts as a sensor for extracellular calcium during vertebrate left–right determination. Nature 427, 121–128 (2004). The authors identify an LR difference in the levels of extracellular Ca2+ in the chick embryo, which are translated by the Notch signalling pathways into asymmetries at the level of NODAL expression. This paper is an example of how mathematical modelling is becoming increasingly important in developmental biology to understand how stable domains of gene expression are produced.
Adachi, H. et al. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes Dev. 13, 1589–1600 (1999).
Norris, D. P. & Robertson, E. J. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev. 13, 1575–1588 (1999).
Brennan, J., Norris, D. P. & Robertson, E. J. Nodal activity in the node governs left–right asymmetry. Genes Dev. 16, 2339–2344 (2002).
Saijoh, Y., Oki, S., Ohishi, S. & Hamada, H. Left–right patterning of the mouse lateral plate requires nodal produced in the node. Dev. Biol. 256, 160–172 (2003).
Krebs, L. T. et al. Notch signaling regulates left–right asymmetry determination by inducing Nodal expression. Genes Dev. 17, 1207–1212 (2003).
Raya, A. et al. Notch activity induces Nodal expression and mediates the establishment of left–right asymmetry in vertebrate embryos. Genes Dev. 17, 1213–1218 (2003).
Nonaka, S. et al. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 (1998).
Okada, Y. et al. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol. Cell 4, 459–468 (1999). Together with reference 32, this paper characterizes the nodal monocilia rotation and nodal flow in the mouse embryo, and their correlation with normal LR patterning.
Takeda, S. et al. Left–right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J. Cell Biol. 145, 825–836 (1999).
Supp, D. M. et al. Targeted deletion of the ATP binding domain of left–right dynein confirms its role in specifying development of left–right asymmetries. Development 126, 5495–5504 (1999).
Marszalek, J. R., Ruiz-Lozano, P., Roberts, E., Chien, K. R. & Goldstein, L. S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl Acad. Sci. USA 96, 5043–5048 (1999).
Murcia, N. S. et al. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left–right axis determination. Development 127, 2347–2355 (2000).
Kobayashi, Y. et al. Hydrocephalus, situs inversus, chronic sinusitis, and male infertility in DNA polymerase λ-deficient mice: possible implication for the pathogenesis of immotile cilia syndrome. Mol. Cell. Biol. 22, 2769–2776 (2002).
Ibañez-Tallon, I., Gorokhova, S. & Heintz, N. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum. Mol. Genet. 11, 715–721 (2002).
Huangfu, D. et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 (2003). The authors uncover a link between intraflagellar transport proteins, primary cilia formation and HH signalling in the mouse embryo.
Rana, A. A. et al. Targeted deletion of the novel cytoplasmic dynein mD2LIC disrupts the embryonic organiser, formation of the body axes and specification of ventral cell fates. Development 131, 4999–5007 (2004).
Bonnafe, E. et al. The transcription factor RFX3 directs nodal cilium development and left–right asymmetry specification. Mol. Cell. Biol. 24, 4417–4427 (2004).
Huangfu, D. & Anderson, K. V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA 102, 11325–11330 (2005).
Nonaka, S., Shiratori, H., Saijoh, Y. & Hamada, H. Determination of left–right patterning of the mouse embryo by artificial nodal flow. Nature 418, 96–99 (2002).
McGrath, J., Somlo, S., Makova, S., Tian, X. & Brueckner, M. Two populations of node monocilia initiate left–right asymmetry in the mouse. Cell 114, 61–73 (2003). The authors uncover an LR difference in the levels of intracellular Ca2+ in the mouse embryo and provide experimental evidence for the two-cilia model.
Cartwright, J. H., Piro, O. & Tuval, I. Fluid-dynamical basis of the embryonic development of left–right asymmetry in vertebrates. Proc. Natl Acad. Sci. USA 101, 7234–7239 (2004).
Nonaka, S. et al. De novo formation of left–right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 3, e268 (2005).
Okada, Y., Takeda, S., Tanaka, Y., Izpisua Belmonte, J. C. & Hirokawa, N. Mechanism of ciliated organ flow: a conserved symmetry breaking event in left–right axis determination. Cell 121, 633–644 (2005).
Buceta, J. et al. Nodal cilia dynamics and the specification of the left/right axis in early vertebrate embryo development. Biophys. J. 89, 2199–2209 (2005). Together with references 47 and 48, this paper shows that the posterior tilting of nodal monocilia, coupled with non-planar beating cilia dynamics, results in a robust leftward fluid flow.
Kramer-Zucker, A. G. et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132, 1907–1921 (2005).
Essner, J. J., Amack, J. D., Nyholm, M. K., Harris, E. B. & Yost, H. J. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left–right development of the brain, heart and gut. Development 132, 1247–1260 (2005).
Amack, J. D. & Yost, H. J. The T box transcription factor no tail in ciliated cells controls zebrafish left–right asymmetry. Curr. Biol. 14, 685–690 (2004).
Meyers, E. N. & Martin, G. R. Differences in left–right axis pathways in mouse and chick: functions of FGF8 and SHH. Science 285, 403–406 (1999).
Rankin, C. T., Bunton, T., Lawler, A. M. & Lee, S. J. Regulation of left–right patterning in mice by growth/differentiation factor-1. Nature Genet. 24, 262–265 (2000).
Tanaka, Y., Okada, Y. & Hirokawa, N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left–right determination. Nature 435, 172–177 (2005). The authors characterize an NVP mechanism of morphogen transport by the nodal flow in the mouse.
Tsukui, T. et al. Multiple left–right asymmetry defects in Shh−/− mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc. Natl Acad. Sci. USA 96, 11376–11381 (1999).
Zhang, X. M., Ramalho-Santos, M. & McMahon, A. P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell 106, 781–792 (2001). This study analysed the LR phenotypes of mouse Smo mutants and uncovered key roles of HH signalling during mouse LR asymmetrical patterning.
Izraeli, S. et al. The SIL gene is required for mouse embryonic axial development and left–right specification. Nature 399, 691–694 (1999).
Lohr, J. L., Danos, M. C., Groth, T. W. & Yost, H. J. Maintenance of asymmetric nodal expression in Xenopus laevis. Dev. Genet. 23, 194–202 (1998).
Levin, M. & Mercola, M. Evolutionary conservation of mechanisms upstream of asymmetric Nodal expression: reconciling chick and Xenopus. Dev. Genet. 23, 185–193 (1998).
Sampath, K., Cheng, A. M., Frisch, A. & Wright, C. V. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development 124, 3293–3302 (1997).
Schilling, T. F., Concordet, J. P. & Ingham, P. W. Regulation of left–right asymmetries in the zebrafish by Shh and BMP4. Dev. Biol. 210, 277–287 (1999).
Goodrich, L. V., Johnson, R. L., Milenkovic, L., McMahon, J. A. & Scott, M. P. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10, 301–312 (1996).
Collignon, J., Varlet, I. & Robertson, E. J. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381, 155–158 (1996).
Lowe, L. A. et al. Conserved left–right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381, 158–161 (1996). Together with reference 64, this paper demonstrates that left-sided expression of Nodal is crucial for normal LR patterning in the mouse.
Lustig, K. D. et al. A Xenopus nodal-related gene that acts in synergy with noggin to induce complete secondary axis and notochord formation. Development 122, 3275–3282 (1996).
Sampath, K. et al. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature 395, 185–189 (1998).
Long, S., Ahmad, N. & Rebagliati, M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left–right asymmetry. Development 130, 2303–2316 (2003).
Zile, M. H. et al. Retinoid signaling is required to complete the vertebrate cardiac left/right asymmetry pathway. Dev. Biol. 223, 323–338 (2000).
Fischer, A., Viebahn, C. & Blum, M. FGF8 acts as a right determinant during establishment of the left–right axis in the rabbit. Curr. Biol. 12, 1807–1816 (2002).
Schier, A. F. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19, 589–621 (2003).
Raya, A. & Izpisúa Belmonte, J. C. Sequential transfer of left–right information during vertebrate embryo development. Curr. Opin. Genet. Dev. 14, 575–581 (2004).
St Amand, T. R. et al. Cloning and expression pattern of chicken Pitx2: a new component in the SHH signaling pathway controlling embryonic heart looping. Biochem. Biophys. Res. Commun. 247, 100–105 (1998).
Ryan, A. K. et al. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nature 394, 545–551 (1998).
Logan, M., Pagan-Westphal, S. M., Smith, D. M., Paganessi, L. & Tabin, C. J. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left–right asymmetric signals. Cell 94, 307–317 (1998).
Piedra, M. E., Icardo, J. M., Albajar, M., Rodriguez-Rey, J. C. & Ros, M. A. Pitx2 participates in the late phase of the pathway controlling left–right asymmetry. Cell 94, 319–324 (1998).
Yoshioka, H. et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left–right asymmetry. Cell 94, 299–305 (1998). Together with references 73–76, this paper characterizes the role of the NODAL-target gene Pitx2 in the LR asymmetrical patterning of vertebrates.
Shiratori, H. et al. Two-step regulation of left–right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7, 137–149 (2001).
Pennekamp, P. et al. The ion channel polycystin-2 is required for left–right axis determination in mice. Curr. Biol. 12, 938–943 (2002).
Constam, D. B. & Robertson, E. J. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development 127, 245–254 (2000).
Boorman, C. J. & Shimeld, S. M. The evolution of left–right asymmetry in chordates. Bioessays 24, 1004–1011 (2002).
Chea, H. K., Wright, C. V. & Swalla, B. J. Nodal signaling and the evolution of deuterostome gastrulation. Dev. Dyn. 234, 269–278 (2005).
Aihara, M. & Amemiya, S. Left–right positioning of the adult rudiment in sea urchin larvae is directed by the right side. Development 128, 4935–4948 (2001).
Dubrulle, J. & Pourquie, O. Coupling segmentation to axis formation. Development 131, 5783–5793 (2004).
Vermot, J. et al. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science 308, 563–566 (2005).
Vermot, J. & Pourquie, O. Retinoic acid coordinates somitogenesis and left–right patterning in vertebrate embryos. Nature 435, 215–220 (2005). Together with references 20 and 85, this paper uncovers a link between the pathways that control LR asymmetrical patterning and bilaterally symmetrical somitogenesis.
Minguillon, C. & Garcia-Fernandez, J. The single amphioxus Mox gene: insights into the functional evolution of Mox genes, somites, and the asymmetry of amphioxus somitogenesis. Dev. Biol. 246, 455–465 (2002).
Hofer, A. M. & Brown, E. M. Extracellular calcium sensing and signalling. Nature Rev. Mol. Cell Biol. 4, 530–538 (2003).
Norris, D. P., Brennan, J., Bikoff, E. K. & Robertson, E. J. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129, 3455–3468 (2002).
Levin, M. & Mercola, M. Gap junctions are involved in the early generation of left–right asymmetry. Dev. Biol. 203, 90–105 (1998).
Levin, M. & Mercola, M. Gap junction-mediated transfer of left–right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node. Development 126, 4703–4714 (1999).
Reaume, A. G. et al. Cardiac malformation in neonatal mice lacking connexin43. Science 267, 1831–1834 (1995).
Iovine, M. K., Higgins, E. P., Hindes, A., Coblitz, B. & Johnson, S. L. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev. Biol. 278, 208–219 (2005).
Echelard, Y. et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417–1430 (1993).
Chiang, C. et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 (1996).
Levin, M., Roberts, D. J., Holmes, L. B. & Tabin, C. Laterality defects in conjoined twins. Nature 384, 321 (1996).
Meno, C. et al. lefty-1 is required for left–right determination as a regulator of lefty-2 and nodal. Cell 94, 287–297 (1998).
Corbit, K. C. et al. Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 (2005).
van den Heuvel, M. & Ingham, P. W. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 382, 547–551 (1996).
Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M. & Hooper, J. E. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86, 221–232 (1996).
Psychoyos, D. & Stern, C. D. Restoration of the organizer after radical ablation of Hensen's node and the anterior primitive streak in the chick embryo. Development 122, 3263–3273 (1996).
Yuan, S. & Schoenwolf, G. C. Reconstitution of the organizer is both sufficient and required to re-establish a fully patterned body plan in avian embryos. Development 126, 2461–2473 (1999).
Pagan-Westphal, S. M. & Tabin, C. J. The transfer of left–right positional information during chick embryogenesis. Cell 93, 25–35 (1998).
Acknowledgements
The authors thank C. Rodríguez-Esteban for providing chick embryo pictures, S. Shimeld and A. Nishino for communicating results prior to publication, J. Simon for excellent artwork, C. Stern and all the members of JCIB laboratory for fruitful discussions, and M.-F. Schwarz for help in the preparation of this manuscript. A.R. was partially supported by a postdoctoral fellowship from Fundación Inbiomed, Spain. The research on left–right asymmetry in our laboratory is supported by the US National Institutes of Health, the Human Frontier Science Program, and the G. Harold and Leila Y. Mathers Charitable Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Embryo node
-
A transient structure located at the anterior tip of the primitive streak in embryos of amniotes (birds, reptiles and mammals); also known as Hensen's node in birds and mammals. The embryo node functions as the gastrula organizer and is therefore functionally equivalent to the dorsal lip of the blastopore (Spemann's organizer) in amphibians and the shield of teleost fishes.
- Lateral plate mesoderm
-
The most lateral region of mesoderm in the neurula-stage vertebrate embryo. Among other structures, it gives rise to the heart, blood vessels, blood cells of the circulatory system, the lining of the body cavities, and all the mesodermal components of the limbs other than muscle.
- Primitive streak
-
A transitory embryonic structure, which is present as a strip of cells, that pre-figures the anterior–posterior axis of the embryo. During gastrulation, embryonic cells progress through the streak.
- Deuterostomes
-
A taxon of animals that belong to the Bilateria. They are characterized by having a 'second mouth' (giving them their name) — that is, during embryo development, the blastopore becomes the anus, whereas the mouth forms in a secondary anterior location. Deuterostomes are divided into two major clades: Ambulacraria (which includes echinoderms and hemichordates) and Chordata (which includes vertebrates, urochordates and cephalochordates).
- Morpholinos
-
Morpholino-modified antisense oligonucleotides (generally known as 'morpholinos') are reagents that are widely used to knockdown gene function in zebrafish by pairing to complementary sequences in gene transcripts and blocking their translation or splicing.
- Posterior notochordal plate
-
The posterior part of the notochordal plate that lies adjacent to the node. It is a flattened, grooved plate, which originates as a result of the fusion and subsequent disappearance of the floor of the notochordal process with the underlying endoderm. The notochordal plate eventually folds inwards to give rise to the notochord.
- Midline barrier
-
A physical and/or molecular barrier that separates the right and left halves of the vertebrate embryo so that the action of long-range side-specific signals does not affect the other side. Physical elements of the barrier are exemplified by the midline derivatives of the node: the notochord and the floorplate. The best-understood molecular component of the midline barrier is the divergent TGFB signal LEFTY1.
- Somitogenesis
-
The process of metameric segmentation of chordate embryos. In this process, paired blocks of paraxial mesoderm (somites) are specified and segmented following a stereotypical species-specific sequence. In vertebrates, somites form in a bilaterally symmetrical fashion and give rise to bilaterally symmetrical structures, such as the skeletal muscles, the axial skeleton and parts of the dermis.
- Rudiment
-
A structure that is present in the larvae of sea urchins that gives rise to most of the adult tissues. The process of rudiment specification is left–right asymmetrical, originating from the left coelomic pouch and its adjacent lateral ectoderm.
- Floorplate
-
Ventral region of the early neural tube of vertebrate embryos. The medial part of the floorplate is formed by cells that originate in the node or organizer, which induces floorplate-like characteristics in the neural ectoderm-derived cells of the lateral floorplate. The floorplate has important roles during ventral nervous system patterning, including the specification of motor neurons and interneurons, and the differentiation of oligodendrocytes.
Rights and permissions
About this article
Cite this article
Raya, Á., Belmonte, J. Left–right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet 7, 283–293 (2006). https://doi.org/10.1038/nrg1830
Issue Date:
DOI: https://doi.org/10.1038/nrg1830
This article is cited by
-
Situs Deconstructed
Pediatric Cardiology (2023)
-
Genetic architecture of laterality defects revealed by whole exome sequencing
European Journal of Human Genetics (2019)
-
A right-handed signalling pathway drives heart looping in vertebrates
Nature (2017)
-
Diaphanous gene mutation affects spiral cleavage and chirality in snails
Scientific Reports (2016)
-
The Nodal signaling pathway controls left-right asymmetric development in amphioxus
EvoDevo (2015)