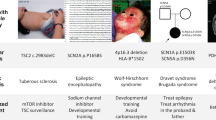
Key Points
-
Target selection and validation is a crucial first step in developing a drug against a given disease. Human genetic analysis is an important model for the identification and/or validation of novel therapeutic targets.
-
Monogenic disorders, involving rare high-penetrance mutations, comprise the most easily interpretable component of experimental human genetics. Because mutations alter the level of activity of gene products, they can be thought of as surrogates for perfectly targeted drugs, the job of which is to antagonize or agonize a given gene product.
-
Phenotypes of some monogenic disease states are of immediate relevance to major therapeutic programmes in the biotechnology and pharmaceutical sectors. The genetics of anti-disease states, such as low blood glucose or low plasma cholesterol, provides a novel approach to new target identification.
-
Among monogenic disorders that have understood molecular bases, many of the causal genes are 'druggable' using classical pharmaceutical methodologies. New chemistries will increase the fraction of the genome that is accessible to small molecular therapeutics.
-
Of the estimated 25,000 genes in the human genome repertoire, only approximately 1,500 have known monogenic disorders that are associated with high-penetrance mutations. Therefore, most of the genome remains to be characterized in terms of monogenic phenotypes. Some of these phenotypes are likely to be relevant to therapeutic developments for medically and commercially important diseases.
-
Ascertainment and molecular characterization of the complete monogenic human genome will require dedicated efforts in clinical genetics, bringing together medical descriptions of known and novel phenotypes with improved technological methods for gene discovery.
Abstract
The decrease in new drug applications and approvals over the past several years results from an underlying crisis in drug target identification and validation. Model organisms are being used to address this problem, in combination with novel approaches such as the International HapMap Project. What has been underappreciated is that discovery of new drug targets can also be revived by traditional Mendelian genetics. A large fraction of the human gene repertoire remains phenotypically uncharacterized, and is likely to encode many unanticipated and novel phenotypes that will be of interest to pharmaceutical and biotechnological drug developers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Frantz, S. FDA publishes analysis of the pipeline problem. Nature Rev. Drug Discov. 3, 379 (2004).
Sams-Dodd, F. Target-based drug discovery: is something wrong? Drug Discov. Today 10, 139–147 (2005). The most recent of several reviews that discuss the challenges facing drug development pipelines and propose various genomic approaches to improve target selection and validation.
Butcher, S. P. Target discovery and validation in the post-genomic era. Neurochem. Res. 28, 367–371 (2003).
Hardy, L. W. & Peet, N. P. The multiple orthogonal tools approach to define molecular causation in the validation of druggable targets. Drug Discov. Today 9, 117–126 (2004).
Lindsay, M. A. Target discovery. Nature Rev. Drug Discov. 2, 831–838 (2003).
Wienholds, E. et al. Efficient target-selected mutagenesis in zebrafish. Genome Res. 13, 2700–2707 (2003).
Mullins, M. C., Hammerschmidt, M., Haffter, P. & Nusslein-Volhard, C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 4, 189–202 (1994).
Quwailid, M. M. et al. A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm. Genome. 15, 585–591 (2004).
Clark, A. T. et al. Implementing large-scale ENU mutagenesis screens in North America. Genetica 122, 51–64 (2004).
Rastan, S. et al. Towards a mutant map of the mouse — new models of neurological, behavioural, deafness, bone, renal and blood disorders. Genetica 122, 47–49 (2004).
Zambrowicz, B. P. et al. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392, 608–611 (1998).
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Cardon, L. R. & Abecasis, G. R. Using haplotype blocks to map human complex trait loci. Trends Genet. 19, 135–140 (2003). A thoughtful discussion of the methodological basis for the HapMap project, its potential, technical challenges and practical guidelines for experimental analysis of complex human traits including common diseases.
Freimer, N. & Sabatti, C. The human phenome project. Nature Genet. 34, 15–21 (2003).
Allen, M. J. & Carrey, A. H. Target identification and validation through genetics. Drug Discov. Today Targets 3, 183–190 (2004).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nature Rev. Genet. 6, 95–108 (2005).
Leroi, A. M. Mutants (Viking Penguin Books, New York, 2003).
Brunner, H. G. & van Driel, M. A. From syndrome families to functional genomics. Nature Rev. Genet. 5, 545–551 (2004). An informative discussion of rare monogenic human disorders and their utility in dissecting basic physiological mechanisms of human development and disease.
Donnai, D. & Read, A. P. How clinicians add to knowledge of development. Lancet 362, 477–484 (2003).
Samuels, M. E. & Dube, M. P. Encyclopedia of Genetics, Genomics, Proteomics, and Bioinformatics Genetics Vol. (John Wiley & Sons, 2005).
Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nature Genet. 33, S228–S237 (2003). An authoritative presentation of methods in human genetic discovery by key exponents in the field, with an historical discussion of past, present and future approaches in monogenic and complex trait analysis
Craig, D. W. & Stephan, D. A. Applications of whole-genome high-density SNP genotyping. Expert Rev. Mol. Diagn. 5, 159–170 (2005).
Horikawa, Y. et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nature Genet. 26, 163–175 (2000).
Song, Y., Niu, T., Manson, J. E., Kwiatkowski, D. J. & Liu, S. Are variants in the CAPN10 gene related to risk of type 2 diabetes? A quantitative assessment of population and family-based association studies. Am. J. Hum. Genet. 74, 208–222 (2004).
Weedon, M. N. et al. Meta-analysis and a large association study confirm a role for calpain-10 variation in type 2 diabetes susceptibility. Am. J. Hum. Genet. 73, 1208–1212 (2003).
Fingerlin, T. E. et al. Variation in three single nucleotide polymorphisms in the calpain-10 gene not associated with type 2 diabetes in a large Finnish cohort. Diabetes 51, 1644–1648 (2002).
Rokman, A. et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 70, 1299–1304 (2002).
Carpten, J. et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nature Genet. 30, 181–184 (2002).
Tavtigian, S. V. et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nature Genet. 27, 172–180 (2001).
Wang, L. et al. Role of HPC2/ELAC2 in hereditary prostate cancer. Cancer Res. 61, 6494–6499 (2001).
Camp, N. J. & Tavtigian, S. V. Meta-analysis of associations of the Ser217Leu and Ala541Thr variants in ELAC2 (HPC2) and prostate cancer. Am. J. Hum. Genet. 71, 1475–1478 (2002).
Styrkarsdottir, U. et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 1, e69 (2003).
Stefansson, H. et al. Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 71, 877–892 (2002).
Tosato, S., Dazzan, P. & Collier, D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr. Bull. 31, 613–617 (2005).
Gretarsdottir, S. et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nature Genet. 35, 131–138 (2003).
Hageman, G. S. et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl Acad. Sci. USA 102, 7227–7232 (2005). One of several reports of the most consistent mapping of a complex genetic trait through high-density SNP-based linkage disequilibrium analysis (see following references for replication and validation of the results).
Klein, R. J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Edwards, A. O. et al. Complement factor H polymorphism and age-related macular degeneration. Science 308, 421–424 (2005).
Haines, J. L. et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 308, 419–421 (2005).
McKusick, V. A. Online Mendelian Inheritance in Man, OMIM (McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University and National Center for Biotechnology Information, National Library of Medicine, Baltimore and Bethesda, Maryland, 2000).
Southan, C. Has the yo-yo stopped? An assessment of human protein-coding gene number. Proteomics 4, 1712–1726 (2004).
Zambrowicz, B. P. & Sands, A. T. Knockouts model the 100 best-selling drugs — will they model the next 100? Nature Rev. Drug Discov. 2, 38–51 (2003). Phenotypic description of engineered mouse knockouts in gene targets of 100 drugs that are available on the market, documenting the relevance of model-system genetics for human pharmaceutical development.
IMS Health. 2004 year-end U.S. prescription and sales information and commentary. IMS Press Room web site, [online] (2005).
Cohen, M. M. Jr. Persistent hyperinsulinemic hypoglycemia of infancy. Am. J. Med. Genet. A 122, 351–353 (2003).
Austin, M. A., Hutter, C. M., Zimmern, R. L. & Humphries, S. E. Genetic causes of monogenic heterozygous familial hypercholesterolemia: a HuGE prevalence review. Am. J. Epidemiol. 160, 407–420 (2004).
Reihner, E. et al. Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N. Engl. J. Med. 323, 224–228 (1990).
Smith, E. P. et al. Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N. Engl. J. Med. 331, 1056–1061 (1994).
Yen, S. S. Clinical applications of gonadotropin-releasing hormone and gonadotropin-releasing hormone analogs. Fertil. Steril. 39, 257–266 (1983).
Spencer, F. A. et al. Enoxaparin, a low molecular weight heparin, inhibits platelet-dependent prothrombinase assembly and activity by factor-Xa neutralization. J. Thromb. Thrombolysis 9, 223–228 (2000).
Storey, R. F. The P2Y12 receptor as a therapeutic target in cardiovascular disease. Platelets 12, 197–209 (2001).
Martens, F. M., Visseren, F. L., Lemay, J., de Koning, E. J. & Rabelink, T. J. Metabolic and additional vascular effects of thiazolidinediones. Drugs 62, 1463–1480 (2002).
Hauptman, J. B., Jeunet, F. S. & Hartmann, D. Initial studies in humans with the novel gastrointestinal lipase inhibitor Ro 18–0647 (tetrahydrolipstatin). Am. J. Clin. Nutr. 55, S309–S313 (1992).
Glaspy, J. The impact of epoetin alfa on quality of life during cancer chemotherapy: a fresh look at an old problem. Semin. Hematol. 34, 20–26 (1997).
Mohini, R. Clinical efficacy of recombinant human erythropoietin in hemodialysis patients. Semin. Nephrol. 9, 16–21 (1989).
Kass, R. S., Arena, J. P. & Chin, S. Cellular electrophysiology of amlodipine: probing the cardiac L-type calcium channel. Am. J. Cardiol. 64, 35I–41I; discussion 41I–42I (1989).
Simon, D. B. et al. Gitelman's variant of Bartter's syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na–Cl cotransporter. Nature Genet. 12, 24–30 (1996).
The MK-906 (Finasteride) Study Group. One-year experience in the treatment of benign prostatic hyperplasia with finasteride. J. Androl. 12, 372–375 (1991).
de Grooth, G. J. et al. A review of CETP and its relation to atherosclerosis. J. Lipid Res. 45, 1967–1974 (2004).
Forrester, J. S., Makkar, R. & Shah, P. K. Increasing high-density lipoprotein cholesterol in dyslipidemia by cholesteryl ester transfer protein inhibition: an update for clinicians. Circulation 111, 1847–1854 (2005).
Hirano, K., Yamashita, S. & Matsuzawa, Y. Pros and cons of inhibiting cholesteryl ester transfer protein. Curr. Opin. Lipidol. 11, 589–596 (2000).
Brousseau, M. E. et al. Effects of an inhibitor of cholesteryl ester transfer protein on HDL cholesterol. N. Engl. J. Med. 350, 1505–1515 (2004).
Brooks-Wilson, A. et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nature Genet. 22, 336–345 (1999).
Rust, S. et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nature Genet. 22, 352–355 (1999).
Lawn, R. M. et al. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J. Clin. Invest. 104, R25–R31 (1999).
Singaraja, R. R. et al. Increased ABCA1 activity protects against atherosclerosis. J. Clin. Invest. 110, 35–42 (2002).
Nassar, M. A. et al. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc. Natl Acad. Sci. USA 101, 12706–12711 (2004).
Yang, Y. et al. Mutations in SCN9A, encoding a sodium channel α-subunit, in patients with primary erythermalgia. J. Med. Genet. 41, 171–174 (2004).
Dib-Hajj, S. D. et al. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 128, 1847–1854 (2005).
Drenth, J. P. et al. SCN9A mutations define primary erythermalgia as a neuropathic disorder of voltage gated sodium channels. J. Invest. Dermatol. 124, 1333–1338 (2005).
Robitaille, J. et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nature Genet. 32, 326–330 (2002).
Robas, N., O'Reilly, M., Katugampola, S. & Fidock, M. Maximizing serendipity: strategies for identifying ligands for orphan G-protein-coupled receptors. Curr. Opin. Pharmacol. 3, 121–126 (2003).
Fredriksson, R. & Schioth, H. B. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 67, 1414–1425 (2005).
Irani, B. G. et al. Progress in the development of melanocortin receptor selective ligands. Curr. Pharm. Des. 10, 3443–3479 (2004).
Kazmierski, W. et al. Recent progress in discovery of small-molecule CCR5 chemokine receptor ligands as HIV-1 inhibitors. Bioorg. Med. Chem. 11, 2663–2676 (2003).
Kondrashov, A. S. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum. Mutat. 21, 12–27 (2003). A meta-analysis of spontaneously arising monogenic human disease states, leading to the most clinically relevant measure of disease-causing mutation rates.
Nachman, M. W. & Crowell, S. L. Estimate of the mutation rate per nucleotide in humans. Genetics 156, 297–304 (2000).
Eyre-Walker, A. & Keightley, P. D. High genomic deleterious mutation rates in hominids. Nature 397, 344–347 (1999).
Zlotogora, J., Shalev, S., Habiballah, H. & Barjes, S. Genetic disorders among Palestinian Arabs: 3. Autosomal recessive disorders in a single village. Am. J. Med. Genet. 92, 343–345 (2000).
de la Chapelle, A. & Wright, F. A. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc. Natl Acad. Sci. USA 95, 12416–12423 (1998). Discussion of methodological approaches to human genetic analysis in founder populations, which focused on Finland as one of the best-studied examples.
Scriver, C. R. Human genetics: lessons from Quebec populations. Annu. Rev. Genomics Hum. Genet. 2, 69–101 (2001).
Rahman, P. et al. The Newfoundland population: a unique resource for genetic investigation of complex diseases. Hum. Mol. Genet. 12, R167–R172 (2003).
Kondo, H., Hayashi, H., Oshima, K., Tahira, T. & Hayashi, K. Frizzled 4 gene (FZD4) mutations in patients with familial exudative vitreoretinopathy with variable expressivity. Br. J. Ophthalmol. 87, 1291–1295 (2003).
Omoto, S., Hayashi, T., Kitahara, K., Takeuchi, T. & Ueoka, Y. Autosomal dominant familial exudative vitreoretinopathy in two Japanese families with FZD4 mutations (H69Y and C181R). Ophthalmic Genet. 25, 81–90 (2004).
Toomes, C. et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am. J. Hum. Genet. 74, 721–730 (2004).
Toomes, C. et al. Spectrum and frequency of FZD4 mutations in familial exudative vitreoretinopathy. Invest. Ophthalmol. Vis. Sci. 45, 2083–2090 (2004).
Yoshida, S. et al. Novel mutation in FZD4 gene in a Japanese pedigree with familial exudative vitreoretinopathy. Am. J. Ophthalmol. 138, 670–671 (2004).
Qin, M. et al. Complexity of the genotype–phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum. Mutat. 26, 104–112 (2005).
Wilson, D. J. et al. A World Wide Web site for low-density lipoprotein receptor gene mutations in familial hypercholesterolemia: sequence-based, tabular, and direct submission data handling. Am. J. Cardiol. 81, 1509–1511 (1998).
Bobadilla, J. L., Macek, M. Jr, Fine, J. P. & Farrell, P. M. Cystic fibrosis: a worldwide analysis of CFTR mutations — correlation with incidence data and application to screening. Hum. Mutat. 19, 575–606 (2002).
Hurst, L. D. & Smith, N. G. Do essential genes evolve slowly? Curr. Biol. 9, 747–750 (1999).
Zambrowicz, B. & Sands, A. T. Modelling drug action in the mouse with knockouts and RNA interference. Drug Discov. Today 3, 198–207 (2004).
Gardner, R. M. & Sutherland, G. R. Chromosome Abnormalities and Genetic Counselling (Oxford Univ. Press, New York, 2004).
Madan, M., Berkowitz, S. D. & Tcheng, J. E. Glycoprotein IIb/IIIa integrin blockade. Circulation 98, 2629–2635 (1998).
Workman, P. New drug targets for genomic cancer therapy: successes, limitations, opportunities and future challenges. Curr. Cancer Drug Targets 1, 33–47 (2001).
Emens, L. A. Trastuzumab: targeted therapy for the management of HER-2/neu-overexpressing metastatic breast cancer. Am. J. Ther. 12, 243–253 (2005).
Berg, T. Modulation of protein-protein interactions with small organic molecules. Angew Chem. Int. Ed. Engl. 42, 2462–2481 (2003).
Arkin, M. R. & Wells, J. A. Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nature Rev. Drug Discov. 3, 301–317 (2004).
Pagliaro, L. et al. Emerging classes of protein–protein interaction inhibitors and new tools for their development. Curr. Opin. Chem. Biol. 8, 442–449 (2004).
Zhao, L. & Chmielewski, J. Inhibiting protein–protein interactions using designed molecules. Curr. Opin. Struct. Biol. 15, 31–34 (2005).
Nishimura, D. Y. et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet–Biedl syndrome gene. Am. J. Hum. Genet. 77, 1021–1033 (2005).
Peltonen, L., Palotie, A. & Lange, K. Use of population isolates for mapping complex traits. Nature Rev. Genet. 1, 182–190 (2000).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Goldstein, D. B., Tate, S. K. & Sisodiya, S. M. Pharmacogenetics goes genomic. Nature Rev. Genet. 4, 937–947 (2003).
Hopkins, A. L. & Groom, C. R. The druggable genome. Nature Rev. Drug Discov. 1, 727–730 (2002). The most-often cited reference for the component of the human genome that encodes classically druggable protein products, which is based on functional domains found in targets of drugs already on the market.
Orth, A. P., Batalov, S., Perrone, M. & Chanda, S. K. The promise of genomics to identify novel therapeutic targets. Expert Opin. Ther. Targets 8, 587–596 (2004). Follow-up reference that updates the set of druggable gene products of the human genome.
Al-Shahrour, F., Diaz-Uriarte, R. & Dopazo, J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20, 578–580 (2004). A simple but powerful web application that can be used to extract Gene Ontology (GO) terms that are differentially represented in sets of genes within the context of genome-scale experiments; part of the Babelomics suite.
Acknowledgements
Thanks to V. McKusick and J. Amberger for data from the Online Mendelian Inheritance in Man. Thanks to M. Ludman, S. Dyack and D. Skidmore for valuable discussions of clinical genetics phenotypes. Thanks to P. Goldberg, R. Sherrington, S. Pimstone for discussions of therapeutic targets. Thanks to S. Chanda for logistic assistance. R.R.B. is supported by a Michael Smith Foundation for Health Research (MSFHR) Research Unit Infrastructure award. M.E.S. is supported by the IWK Health Centre, Capital District Health Authority and Dalhousie University Faculty of Medicine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Glossary
- Haplotype mapping
-
A technique that involves the use of combinations of 'common' DNA polymorphisms to find blocks of association with phenotypic traits.
- Humanized antibodies
-
Antibodies in which only the parts of antibody variable regions that mediate the contact to antigens have been grafted onto a human antibody framework by means of genetic engineering techniques.
- Peptidomimetics
-
Compounds that are derived from peptides and proteins by structural modification using, for example, unnatural amino acids, conformational restraints, isosteric replacement and cyclization.
- Pleiotropic
-
The phenomenon in which a single gene is responsible for several distinct and seemingly unrelated phenotypic effects.
- Minor allele frequency
-
The frequency of the less common allele of a polymorphic locus. It has a value that lies between 0 and 0.5, and can vary between populations.
- Endometriosis
-
A common medical condition in which the tissue lining the uterus (the endometrium) is found outside the uterus, typically affecting other organs in the pelvis.
- Founder populations
-
Populations that that have been derived from a limited pool of individuals within the last 100 or fewer generations.
- Compound heterozygosity
-
A situation in which an inidividual is heterozygous for two different mutations at the same locus.
- Proband
-
A subject that is ascertained on the basis of phenotype; they are often used to identify affected families for genetic studies.
Rights and permissions
About this article
Cite this article
Brinkman, R., Dubé, MP., Rouleau, G. et al. Human monogenic disorders — a source of novel drug targets. Nat Rev Genet 7, 249–260 (2006). https://doi.org/10.1038/nrg1828
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg1828
This article is cited by
-
Discovery of mutations for Mendelian disorders
Human Genetics (2016)
-
A comparative study of disease genes and drug targets in the human protein interactome
BMC Bioinformatics (2015)
-
Genetic similarity between cancers and comorbid Mendelian diseases identifies candidate driver genes
Nature Communications (2015)
-
Rational drug repositioning by medical genetics
Nature Biotechnology (2013)
-
Loss of Siglec-14 reduces the risk of chronic obstructive pulmonary disease exacerbation
Cellular and Molecular Life Sciences (2013)