Key Points
-
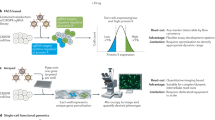
High-throughput screens, in which each gene of an organism is systematically perturbed, are now routine in yeast and are becoming feasible in other organisms.
-
Before conducting a genome-wide screen, the list of gene sequences to be targeted must be compiled and a set of gene-perturbing reagents or strains must be constructed.
-
Gene-perturbing strategies include: homologous recombination and random insertional mutagenesis to delete or mutate genes at the DNA level, and RNA interference to reduce the mRNA levels of a gene.
-
Any observable phenotype can be screened using genome-wide collections of reagents or organisms, subject to practical limitations.
-
Examples of phenotypes that have been screened so far include cell growth and proliferation, classical morphological defects and reporter-gene activity.
-
Using automated microscopy and/or automated image analysis, visual phenotypes can also be screened in a high-throughput manner.
-
Genome-wide phenotypic screens generate large, high-quality data sets, which can be used immediately to identify genes that are involved in a particular process.
-
These genome-wide data sets can also be integrated with existing genome-wide data sets, such as transcriptional profiles and annotated databases, to provide further, broader insights.
-
Future screens will probably examine more complex phenotypes in increasingly physiological contexts.
Abstract
By using genome information to create tools for perturbing gene function, it is now possible to undertake systematic genome-wide functional screens that examine the contribution of every gene to a biological process. The directed nature of these experiments contrasts with traditional methods, in which random mutations are induced and the resulting mutants are screened for various phenotypes. The first genome-wide functional screens in Caenorhabditis elegans and Drosophila melanogaster have recently been published, and screens in human cells will soon follow. These high-throughput techniques promise the rapid annotation of genomes with high-quality information about the biological function of each gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Marcotte, E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. A combined algorithm for genome-wide prediction of protein function. Nature 402, 83–86 (1999).
Grunenfelder, B. & Winzeler, E. A. Treasures and traps in genome-wide data sets: case examples from yeast. Nature Rev. Genet. 3, 653–661 (2002).
von Mering, C. et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 (2002).
Hammerle, M. et al. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 25000–25005 (1998).
Regelmann, J. et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14, 1652–1663 (2003).
Snyder, M. & Gerstein, M. Genomics. Defining genes in the genomics era. Science 300, 258–260 (2003).
Misra, S. et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3, RESEARCH0083 (2002).
Stein, L. Genome annotation: from sequence to biology. Nature Rev. Genet. 2, 493–503 (2001).
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424, 797–801 (2003).
Heo, W. D. & Meyer, T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell 113, 315–328 (2003).
Goshima, G. & Vale, R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016 (2003).
Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 (2003).
Adams, M. D. & Sekelsky, J. J. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nature Rev. Genet. 3, 189–198 (2002).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). Describes the construction of ∼6,000 yeast deletion strains, and a screen for proliferation under various growth conditions.
Hannon, G. J. RNA interference. Nature 418, 244–251 (2002).
Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. Killing the messenger: short RNAs that silence gene expression. Nature Rev. Mol. Cell Biol. 4, 457–467 (2003).
Shi, Y. Mammalian RNAi for the masses. Trends Genet. 19, 9–12 (2003).
McManus, M. T. & Sharp, P. A. Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet 3, 737–747 (2002).
Kamath, R. S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003).
Hutvagner, G. & Zamore, P. D. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12, 225–232 (2002).
Tavernarakis, N., Wang, S. L., Dorovkov, M., Ryazanov, A. & Driscoll, M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nature Genet 24, 180–183 (2000).
Kanemaki, M., Sanchez-Diaz, A., Gambus, A. & Labib, K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423, 720–724 (2003).
Geyer, C. R., Colman-Lerner, A. & Brent, R. 'Mutagenesis' by peptide aptamers identifies genetic network members and pathway connections. Proc. Natl Acad. Sci. USA 96, 8567–8572 (1999).
Bishop, A. C., Buzko, O. & Shokat, K. M. Magic bullets for protein kinases. Trends Cell Biol. 11, 167–172 (2001).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Steinmetz, L. M. et al. Systematic screen for human disease genes in yeast. Nature Genet. 31, 400–404 (2002).
Ross-Macdonald, P. et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413–418 (1999).
Deutschbauer, A. M., Williams, R. M., Chu, A. M. & Davis, R. W. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 99, 15530–15535 (2002).
Dimmer, K. S. et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 (2002).
Gu, Z. et al. Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66 (2003).
Birrell, G. W., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl Acad. Sci. USA 98, 12608–12613 (2001).
Bennett, C. B. et al. Genes required for ionizing radiation resistance in yeast. Nature Genet. 29, 426–434 (2001).
Game, J. C. et al. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 160, 14–24 (2003).
Hanway, D. et al. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl Acad. Sci. USA 99, 10605–10610 (2002).
Bianchi, M. M. et al. Large-scale phenotypic analysis reveals identical contributions to cell functions of known and unknown yeast genes. Yeast 18, 1397–1412 (2001).
Chan, T. F., Carvalho, J., Riles, L. & Zheng, X. F. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl Acad. Sci. USA 97, 13227–13232 (2000).
Butcher, R. A. & Schreiber, S. L. A small molecule suppressor of FK506 that targets the mitochondria and modulates ionic balance in Saccharomyces cerevisiae. Chem. Biol. 10, 521–531 (2003).
Page, N. et al. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163, 875–894 (2003).
Zewail, A. et al. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl Acad. Sci. USA 100, 3345–3350 (2003).
Chang, M., Bellaoui, M., Boone, C. & Brown, G. W. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA 99, 16934–16939 (2002).
Desmoucelles, C., Pinson, B., Saint-Marc, C. & Daignan-Fornier, B. Screening the yeast 'disruptome' for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277, 27036–27044 (2002).
Gupta, S. S. et al. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J. Biol. Chem. 278, 28831–28839 (2003).
Anderson, J. B. et al. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 163, 1287–1298 (2003).
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). Describes the construction of a genome-wide collection of feedable, bacterially expressed dsRNA for C. elegans , and screening for classical phenotypes. This is the first genome-wide RNAi screen to be carried out in a multicellular organism.
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000).
Maeda, I., Kohara, Y., Yamamoto, M. & Sugimoto, A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11, 171–176 (2001).
Simmer, F. et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveall novel gene functions. PLOS Biology 1, E12 (2003).
Lee, S. S. et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature Genet. 33, 40–48 (2003).
Aza-Blanc, P. et al. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12, 627–637 (2003).
Ooi, S. L., Shoemaker, D. D. & Boeke, J. D. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294, 2552–2556 (2001).
Tong, A. H. et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 (2001). Describes the first use of the synthetic genetic array strategy, in which a mutant of interest is crossed with all yeast deletion strains, allowing a genetic network to be constructed.
Krogan, N. J. et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 23, 4207–4218 (2003).
Huang, D., Moffat, J. & Andrews, B. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell Biol. 22, 5076–5088 (2002).
Kroll, E. S., Hyland, K. M., Hieter, P. & Li, J. J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143, 95–102 (1996).
Hartman, J. L., Garvik, B. & Hartwell, L. Principles for the buffering of genetic variation. Science 291, 1001–1004 (2001).
Zhang, J. et al. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr. Biol. 12, 1992–2001 (2002).
Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J. & Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 (2002).
Wilson, W. A., Wang, Z. & Roach, P. J. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol. Cell Proteomics 1, 232–242 (2002).
Bonangelino, C. J., Chavez, E. M. & Bonifacino, J. S. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486–2501 (2002).
Felder, T. et al. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1, 799–810 (2002).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). Reports the construction of ∼6,000 yeast strains that contain TAP epitope-tagged genes in their natural genomic location. Protein expression was detected by Western blot in ∼4,000 strains.
Lum, L. et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039–2045 (2003). Reports the screening by dsRNA against ∼43% of the Drosophila genome using a reporter assay.
Chanda, S. K. et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc. Natl Acad. Sci. USA 100, 12153–12158 (2003).
Iourgenko, V. et al. Identification of a family of cAMP response element-binding protein co-activators by genome-scale functional analysis in mammalian cells. Proc. Natl Acad. Sci. USA 100, 12147–12152 (2003).
Conkright, M. D. et al. TORCs: transducers of regulated CREB activity. Mol. Cell 12, 413–423 (2003).
Pothof, J. et al. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 17, 443–448 (2003).
Vastenhouw, N. L. et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13, 1311–1316 (2003).
Ni, L. & Snyder, M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147–2170 (2001).
Chen, C. et al. An integrated functional genomics screening program reveals a role for BMP-9 in glucose homeostasis. Nature Biotechnol. 21, 294–301 (2003).
Fiscella, M. et al. TIP, a T-cell factor identified using high-throughput screening increases survival in a graft-versus-host disease model. Nature Biotechnol. 21, 302–307 (2003).
Ziauddin, J. & Sabatini, D. M. Microarrays of cells expressing defined cDNAs. Nature 411, 107–110 (2001).
Mousses, S. et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 13, 2341–2347 (2003).
Kumar, R., Conklin, D. S. & Mittal, V. High-throughput selection of effective RNAi probes for gene silencing. Genome Res. 13, 2333–2340 (2003).
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 4, 67–73 (1999).
Editorial. Whither RNAi? Nature Cell Biol. 5, 489–490 (2003).
Le Bot, N., Tsai, M. C., Andrews, R. K. & Ahringer, J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13, 1499–1505 (2003).
Csete, M. E. & Doyle, J. C. Reverse engineering of biological complexity. Science 295, 1664–1669 (2002).
Greenspan, R. J. The flexible genome. Nature Rev. Genet. 2, 383–387 (2001).
Bader, G. D. et al. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 13, 344–356 (2003).
Vidal, M. A biological atlas of functional maps. Cell 104, 333–339 (2001).
Michiels, F. et al. Arrayed adenoviral expression libraries for functional screening. Nature Biotechnol. 20, 1154–1157 (2002).
Shinagawa, T. & Ishii, S. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes Dev. 17, 1340–1345 (2003).
Rubinson, D. A. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genet. 33, 401–406 (2003).
Hemann, M. T. et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nature Genet. 33, 396–400 (2003).
Zeng, Y., Wagner, E. J. & Cullen, B. R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9, 1327–1333 (2002).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 21, 635–637 (2003).
Feinberg, E. H. & Hunter, C. P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 (2003).
Gonczy, P. et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336 (2000).
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001).
Swedlow, J. R., Goldberg, I., Brauner, E. & Sorger, P. K. Informatics and quantitative analysis in biological imaging. Science 300, 100–102 (2003).
Issel-Tarver, L. et al. Saccharomyces genome database. Methods Enzymol. 350, 329–346 (2002).
Enyenihi, A. H. & Saunders, W. S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163, 47–54 (2003).
Zettel, M. F. et al. The budding index of Saccharomyces cerevisiae deletion strains identifies genes important for cell cycle progression. FEMS Microbiol. Lett. 223, 253–258 (2003).
Wiederkehr, A., Meier, K. D. & Riezman, H. Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis. Yeast 18, 759–773 (2001).
Kumar, A. et al. Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 (2002).
Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). Reports the construction of ∼6,000 yeast strains that contain GFP-tagged genes in their natural genomic location. The localization of tagged product was detected by fluorescence microscopy in ∼4,000 strains.
Ashrafi, K. et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 (2003).
Kiger, A. et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2, 27 (2003).
Reboul, J. et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nature Genet. 34, 35–41 (2003).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). Insertional mutants in ∼75% of Arabidopsis genes were made, although the entire set was not screened.
Beckers, J. & Hrabe de Angelis, M. Large-scale mutational analysis for the annotation of the mouse genome. Curr. Opin. Chem. Biol. 6, 17–23 (2002).
Acknowledgements
The authors thank D. A. Guertin for helpful discussions, and M. Boutros and N. Perrimon for sharing unpublished work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
ColiBase
Model organism databases
H. sapiens (Human genome resources)
M. musculus (Mouse genome informatics)
M. musculus (Mouse genome server)
S. cerevisiae (Saccharomyces genome database)
FURTHER INFORMATION
Glossary
- TRANSCRIPTIONAL PROFILING
-
The expression of thousands of genes can be measured simultaneously by spotting an array of DNA sequences on a glass slide and hybridizing a cell population's fluorescently labelled mRNA (or reverse-transcribed cDNA) to the slide. The fluorescence intensity of each spot corresponds to the prevalence in the cells of that nucleic acid species.
- YEAST TWO-HYBRID ASSAY
-
One protein is fused to a transcriptional activation domain and the other to a DNA-binding domain, and both fusion proteins are introduced into yeast. Expression of a reporter gene with the appropriate DNA binding sites upstream of the promoter indicates that the two proteins physically interact.
- RNA INTERFERENCE
-
(RNAi). The process by which the introduction or expression within cells of single- or double-stranded RNA leads to the degradation of the encoded mRNA and therefore to gene suppression.
- LAMELLA
-
The dense, actin-rich structure that extends the leading edge of a migrating cell.
- microRNAs
-
Tiny, noncoding RNAs that are probably involved in gene regulation.
- HOMOLOGOUS RECOMBINATION
-
The process by which segments of DNA are exchanged between two DNA duplexes that share high sequence similarity.
- MOLECULAR BAR CODES
-
Short, unique, engineered DNA sequences that are used as tags. For example, the bar code on each yeast deletion strain allows the identity of the strain to be determined by sequencing the code or by hybridizing DNA from the strain onto a microarray.
- S2 CELLS
-
A cell line that is isolated from dissociated Drosophila melanogaster embryos. The cell line is phagocytic, which might contribute to its susceptibility to RNAi.
- INTERFERON RESPONSE
-
A primitive antiviral mechanism that triggers sequence-nonspecific degradation of mRNA and downregulation of cellular protein synthesis.
- DEGRON TAG
-
A degron (degradation) tag attached to a protein of interest specifically targets the protein for rapid proteolysis if the cells are grown at high temperature (37°) and in the presence of an overexpressed ubiquitin-associated protein that recognizes the tag.
- PEPTIDE APTAMER INHIBITOR
-
Synthetic proteins that can bind and inhibit protein function.
- TRAIL
-
A member of the tumour necrosis factor superfamily that preferentially induces apoptosis in tumour cells while leaving normal cells intact.
- NON-HOMOLOGOUS END-JOINING
-
(NHEJ). One of two cellular DNA-repair pathways that are involved in the repair of double-strand breaks.
- SYNTHETIC LETHAL
-
Synthetic interactions are identified if mutations in two separate genes produce a different phenotype from either gene alone, and indicate a functional association between the two genes. Two genes have a synthetic lethal relationship if mutants in either gene are viable but the double mutation is lethal.
- CALCOFLUOR
-
A chemical that binds the chitin-rich bud scars that remain on the cell surface after cytokinesis.
- SYNTHETIC DOSAGE LETHALITY
-
This type of genetic interaction is detected when overexpression of a gene is lethal only if another, normally nonlethal, mutation is present.
- COULTER PARTICLE COUNTER
-
An instrument that measures the size of particles on the basis of changes in the electrical voltage as they pass through an orifice.
- EPISTATIC TESTS
-
These can place genes in the same or different pathways and can establish the order of gene function if they are in a single, linear pathway. Gene A is epistatic to gene B if the phenotype that results from mutation of both genes matches the phenotype that results from gene A alone (and does not match that of gene B alone).
- MITOTIC INDEX
-
The percentage of cells in the mitotic phase of the cell cycle.
- FLUORESCENCE PLATE READERS
-
Instruments that read the fluorescence in each well of a multiwell plate.
- FLUORESCENCE RESONANCE ENERGY TRANSFER
-
(FRET). A phenomenon by which the energy from an excited fluorophore is transferred to an acceptor molecule at short (<100 Å) distances, leading to decreased fluorescence of the donor and increased fluorescence of the acceptor. The efficiency of energy transfer depends strongly on the distance between the donor and acceptor molecules.
- FLUORESCENCE ACTIVATED CELL SORTERS
-
(FACS). The separation of cells or chromosomes by their fluorescence and light-scattering properties, which are measured as the particles flow in a liquid stream as they pass through laser beams. The stream is then broken into droplets, and selected droplets are electrically charged and deflected into collection vessels as they pass through an electric field.
- LENTIVIRUS
-
A type of retrovirus that can transduce overexpression and RNAi-inducing constructs to dividing and non-dividing mammalian cells.
- CELL MICROARRAYS
-
An array of gene-perturbing reagents (such as plasmids plus a transfection reagent) that is spotted onto a glass slide. Cells that are are plated onto the slide and that land on a spot are affected by the reagent.
- Z-FACTOR
-
A measurement that takes into account the dynamic range of the assay (how far apart the positive controls are from the negative controls), as well as data variability (how much variation is seen in the measurements of positive and negative controls).
Rights and permissions
About this article
Cite this article
Carpenter, A., Sabatini, D. Systematic genome-wide screens of gene function. Nat Rev Genet 5, 11–22 (2004). https://doi.org/10.1038/nrg1248
Issue Date:
DOI: https://doi.org/10.1038/nrg1248
This article is cited by
-
Robust normalization and transformation techniques for constructing gene coexpression networks from RNA-seq data
Genome Biology (2022)
-
Shared genetic architecture between schizophrenia and subcortical brain volumes implicates early neurodevelopmental processes and brain development in childhood
Molecular Psychiatry (2022)
-
A genome-wide atlas of co-essential modules assigns function to uncharacterized genes
Nature Genetics (2021)
-
High-throughput genetic screens using CRISPR–Cas9 system
Archives of Pharmacal Research (2018)
-
An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks
BMC Systems Biology (2017)