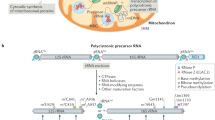
Abstract
Phylogenetic studies of the yeast mitochondrial proteome have shown a complex evolutionary scenario, in which proteins of bacterial origin form complexes with proteins of eukaryotic origin. Exciting new results from whole-genome microarray studies of subcellular mRNA localizations have shown that mRNAs that are of putative bacterial origin are mainly translated on polysomes that are associated with the mitochondrion, whereas those of eukaryotic origin are generally translated on free cytosolic polysomes. Understanding these newly discovered relationships promises insights into old questions about organelle origins and mRNA localization in the eukaryotic cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Saraste, M. Oxidative phosphorylation at the fin de siecle. Science 283, 1488–1492 (1999).
Dillon, A. et al. Rates of behaviour and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002).
Wallace, D. C. Mitochondrial diseases in man and mouse. Science 283, 1482–1487 (1999).
Margulis, L. Symbiosis in Cell Evolution (Freeman, San Francisco, 1981).
Gray, M., Burger, G. & Lang, B. F. Mitochondrial evolution. Science 283, 1476–1481 (1999).
Karlberg, O., Canbäck, B., Kurland, C. G. & Andersson, S. G. E. The dual origin of the yeast mitochondrial proteome. Yeast 17, 170–187 (2000).
Marc et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3, 159–164 (2002).
Farquhar, J., Bao, H. & Thiemans, M. Atmospheric influence on Earth's earliest sulfur cycle. Science 289, 765–769 (2000).
Goodner, B. et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294, 2323–2328 (2001).
Wood, D. W. et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323 (2001).
Kaneko, T. et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7, 331–338 (2000).
DelVecchio, V. G. et al. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl Acad. Sci. USA 99, 443–448 (2002).
Andersson, S. G. E. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140 (1998).
Kurland, C. G. & Andersson, S. G. E. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64, 786–820 (2000).
Canbäck, B., Andersson, S. G. E. & Kurland, C. G. The global phylogeny of glycolytic enzymes. Proc. Natl Acad. Sci. USA 99, 6097–6102 (2002).
Woischnik, M. & Moraes, C. T. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 12, 885–893 (2002).
Hodges, P. E., McKee, A. H., Davis, B. P., Payne, W. E. & Garrels, J. I. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 27, 69–73 (1999).
Kumar, A. Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 (2002).
Marx, S. et al. Structure of the bc1 complex from Seculamonas ecuadoriensis, a jakobid flagellate with an ancestral mitochondrial genome. Mol. Biol. Evol. 20, 145–153 (2003).
Amiri, H., Karlberg, O. & Anderssson, S. G. E. Deep origin of plastid/parasite ATP/ADP translocases. J. Mol. Evol. 56, 137–150 (2003).
Pfanner, N. & Geissler, A. Versatility of the mitochondrial import machinery. Nature Rev. Mol. Cell Biol. 2, 339–349 (2001).
Geissler, A. et al. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111, 507–518 (2002).
Lithgow, T. Targeting of proteins to mitochondria. FEBS Lett. 476, 22–26 (2000).
Gratzer, S., Beilharz, T., Beddoe, T., Henry, M. F. & Lithgow, T. The mitochondrial protein targeting suppressor (mts1) mutation maps to the mRNA-binding domain of Np13p and affects translation on cytoplasmic polysomes. Mol. Microbiol. 35, 1277–1285 (2000).
George, R., Walsh, P., Beddoe, T. & Lithgow, T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 516, 213–216. (2002).
Margeot, A. et al. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 21, 6893–6904 (2002).
Fujiki, M. & Verner, K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast: evidence for co-translational import in vivo. J. Biol. Chem. 268, 1914–1920 (1993).
Steinmetz, L. M. et al. Systematic screen for human disease genes in yeast. Nature Genet. 31, 400–404 (2002).
Chinnery, P. F. & Turnbull, D. M. The epidemiology and treatment of mitochondrial disease. Am. J. Med. Genet. 106, 94–101 (2001).
Yaffe, M. P. The machinery of mitochondria inheritance and behaviour. Science 283, 1493–1497 (1999).
Juan, A. S., Brown, M. D. & Wallace, D. C. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc. Natl Acad. Sci. USA 91, 6206–6210 (1994).
Shoffner, J. M. et al. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA (Lys) mutation. Cell 61, 931–937 (1990).
Goto, Y., Nonaka, I. & Horai, S. A mutation in the tRNA (Leu)(UU) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 (1990).
Bourgeron, T. et al. Mutation of a nuclear succcinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nature Genet. 11, 144–149 (1995).
Loeffen, J. et al. The first nuclear-encoded complex I mutation in a patient with Leigh syndrome. Am. J. Hum. Genet. 63, 1594–1597 (1998).
Tiranti, V. et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63, 1609–1621 (1998).
Graham, B. H. et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nature Genet. 16, 226–234 (1997).
Murdock, D. G., Boone, B. E., Esposito, L. A. & Wallace, D. C. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J. Biol. Chem. 174, 14429–14433 (1999).
Esposito, L. A., Melov, S., Panov, A., Cottrell, B. A. & Wallace, D. C. Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl Acad. Sci. USA 96, 4820–4825 (1999).
Petit, P. X., Susin, S. A., Zamzami, N., Mignotte, B. & Kroemer, G. Mitochondria and programmed cell death: back to the future. FEBS Lett. 396, 7–13 (1996).
Green, D. R. & Reed, J. C. Mitochondria and apoptosis. Science 281, 1309–1312 (1998).
Susin, S. A. et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 387–389 (1999).
Andersson, S. G. E. & Kurland, C. G. Origin of mitochondria and hydrogenosomes. Curr. Opin. Microbiol. 2, 535–541 (1999).
Martin, W. & Mueller, M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998).
Moreira, D. & Lopez-Garcia, P. Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J. Mol. Evol. 47, 517–530 (1998).
Brooks, J. J., Logan, G. A., Buick, R. & Summons, R. E. Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036 (1999).
Kump, L. R., Kasting, J. F. & Barley, M. E. Rise of atmospheric oxygen and the 'upside-down' Archaean mantle. Geol. Geochem. Geophys. 2 (2001).
Catling, D. C., Zahnle, K. J. & McKay, C. P. Biogenic methane, hydrogen escape, and the irreversible oxidation of early Earth. Science 293, 839–843 (2001).
Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. Predicting subcellular localization of proteins based on their N–terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 (2000).
Acknowledgements
The authors thank all the members of the Department of Molecular Evolution at Uppsala University for providing a stimulating atmosphere.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
LocusLink
OMIM
SwissProt
FURTHER INFORMATION
Glossary
- ANAEROBE
-
An organism that can grow in an oxygen-free environment.
- CYANOBACTERIA
-
Photosynthetic bacteria, formerly called blue-green algae.
- ENDOSYMBIONT
-
An organism that grows inside the cell of another organism. The relationship can be either mutualistic (both species benefit) or commensalistic (one species benefits, whereas the other is not affected).
- FACULTATIVE INTRACELLULAR PARASITE
-
A parasite that is capable of growing both inside and outside of the cells of the infected host.
- HORIZONTAL TRANSFER
-
The transfer of genetic material among cells that belong to different strains, species or genera.
- KRUSKAL-WALLIS NON-PARAMETRIC TEST
-
A statistical test that is used to assess whether two (or more) populations have the same mean and distribution.
- MATURASES
-
Enzymes that are involved in removing introns from mRNA.
- POLYSOME
-
(Polyribosome). Many ribosomes that are connected by an mRNA molecule.
- PROTISTS
-
Unicellular eukaryotes including protozoans, slime molds and certain algae.
- VERTICAL DESCENT
-
The transfer of genetic material from an ancestor to its offspring — for example, by cell division.
Rights and permissions
About this article
Cite this article
Karlberg, E., Andersson, S. Mitochondrial gene history and mRNA localization: is there a correlation?. Nat Rev Genet 4, 391–397 (2003). https://doi.org/10.1038/nrg1063
Issue Date:
DOI: https://doi.org/10.1038/nrg1063
This article is cited by
-
Endosymbiont or host: who drove mitochondrial and plastid evolution?
Biology Direct (2011)
-
RNA trafficking in plant cells: targeting of cytosolic mRNAs to the mitochondrial surface
Plant Molecular Biology (2010)
-
Evolution of early eukaryotic cells: genomes, proteomes, and compartments
Photosynthesis Research (2007)
-
mRNA trafficking in fungi
Molecular Genetics and Genomics (2007)
-
The evolution of chronic infection strategies in the α-proteobacteria
Nature Reviews Microbiology (2004)