Key Points
-
Modified nucleotides are present in many RNA species and have multiple roles in the global regulation and fine-tuning of gene expression.
-
Despite growing interest in the discovery and analysis of RNA modification profiles and their dynamics, conventional methods for transcriptome-wide profiling are laborious and time-consuming.
-
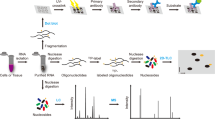
Next-generation sequencing brings new perspectives for global analysis of RNA modification in different cells and tissues under various physiological conditions. Current methods and technologies applied for RNA modification analysis at the transcriptome level are discussed.
-
Most of these techniques use high-throughput DNA sequencing, coupled to the use of specific chemical reagents or specific antibodies to reveal modified RNA nucleotides.
-
An epitranscriptome analysis should be followed by thorough validation of obtained candidate sites by complementary orthogonal approaches.
-
Experimental and bioinformatic approaches and challenges for global analysis of RNA modification are discussed.
Abstract
RNA modifications are emerging players in the field of post-transcriptional regulation of gene expression, and are attracting a comparable degree of research interest to DNA and histone modifications in the field of epigenetics. We now know of more than 150 RNA modifications and the true potential of a few of these is currently emerging as the consequence of a leap in detection technology, principally associated with high-throughput sequencing. This Review outlines the major developments in this field through a structured discussion of detection principles, lays out advantages and drawbacks of new high-throughput methods and presents conventional biophysical identification of modifications as meaningful ways for validation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Saletore, Y. et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012).
Schwartz, S. Cracking the epitranscriptome. RNA 22, 169–174 (2016).
Witkin, K. L. et al. RNA editing, epitranscriptomics, and processing in cancer progression. Cancer Biol. Ther. 16, 21–27 (2015).
Roundtree, I. A. & He, C. RNA epigenetics — chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 30, 46–51 (2016).
Liu, N. & Pan, T. RNA epigenetics. Transl Res. 165, 28–35 (2015).
Spenkuch, F., Motorin, Y. & Helm, M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 11, 1540–1554 (2014).
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways — 2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Helm, M. & Alfonzo, J. D. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 21, 174–185 (2014).
Holley, R. W. et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965).
Ban, E. & Song, E. J. Recent developments and applications of capillary electrophoresis with laser-induced fluorescence detection in biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 929, 180–186 (2013).
Karijolich, J. & Yu, Y. T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Zinshteyn, B. & Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 202–209 (2009).
Yu, B. et al. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 (2005).
Motorin, Y. & Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631 (2011).
Moss, B., Gershowitz, A., Stringer, J. R., Holland, L. E. & Wagner, E. K. 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 23, 234–239 (1977).
Nichols, J. L. & Welder, L. The modified nucleotide constituents of human prostatic cancer cell (MA-160) poly(A)-containing RNA. Biochim. Biophys. Acta 608, 1–18 (1980).
Rottman, F. M., Desrosiers, R. C. & Friderici, K. Nucleotide methylation patterns in eukaryotic mRNA. Prog. Nucleic Acid. Res. Mol. Biol. 19, 21–38 (1976).
Limbach, P. A. & Paulines, M. J. Going global: the new era of mapping modifications in RNA. Wiley Interdiscip. Rev. RNA 8, e1367 (2016).
Song, C. X., Yi, C. & He, C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 30, 1107–1116 (2012).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). This is the first demonstration of dynamics in mRNA modification by enzymatic demethylation.
Ebhardt, H. A. et al. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 37, 2461–2470 (2009).
Findeiss, S., Langenberger, D., Stadler, P. F. & Hoffmann, S. Traces of post-transcriptional RNA modifications in deep sequencing data. Biol. Chem. 392, 305–313 (2011).
Ryvkin, P. et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692 (2013).
Alon, S. et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 22, 1533–1540 (2012).
Dominissini, D., Moshitch-Moshkovitz, S., Amariglio, N. & Rechavi, G. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis 32, 1569–1577 (2011).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015). This is a combination of the cDNA signature concept with machine learning to predict modification sites.
Tserovski, L. et al. High-throughput sequencing for 1-methyladenosine (m1A) mapping in RNA. Methods 107, 110–121 (2016).
Gustafsson, C. & Persson, B. C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180, 359–365 (1998).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Talkish, J., May, G., Lin, Y., Woolford, J. L. Jr & McManus, C. J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20, 713–720 (2014).
Weeks, K. M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20, 295–304 (2010).
Sakurai, M., Yano, T., Kawabata, H., Ueda, H. & Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 6, 733–740 (2010). This is pioneering work on the use of selective chemicals for transcriptome-wide detection of modifications.
Suzuki, T., Ueda, H., Okada, S. & Sakurai, M. Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat. Protoc. 10, 715–732 (2015).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9, e110799 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015). In this paper, a chemical pulldown is combined with thorough validation of pseudouridine in mRNA.
Amort, T. et al. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 10, 1003–1008 (2013).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 (2009).
Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Burgess, A. L., David, R. & Searle, I. R. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 15, 199 (2015).
Militello, K. T., Chen, L. M., Ackerman, S. E., Mandarano, A. H. & Valentine, E. L. A map of 5-methylcytosine residues in Trypanosoma brucei tRNA revealed by sodium bisulfite sequencing. Mol. Biochem. Parasitol. 193, 122–126 (2014).
Schaefer, M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 560, 297–329 (2015). The adaptation of bisulfite sequencing from DNA to RNA has strongly affected the field by pointing out the possibility for transcriptome-wide studies.
Krueger, F., Kreck, B., Franke, A. & Andrews, S. R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 9, 145–151 (2012).
Booth, M. J., Marsico, G., Bachman, M., Beraldi, D. & Balasubramanian, S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6, 435–440 (2014).
Van Haute, L. et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 7, 12039 (2016). This shows the combined application of three different methods for the detection of m5C and derivatives in mitochondrial RNA.
Behm-Ansmant, I., Helm, M. & Motorin, Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J. Nucleic Acids 2011, 408053 (2011).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 (2016).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Zaringhalam, M. & Papavasiliou, F. N. Pseudouridylation meets next-generation sequencing. Methods 107, 63–72 (2016).
Hussain, S., Aleksic, J., Blanco, S., Dietmann, S. & Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14, 215 (2013).
Jeltsch, A. et al. Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA Biol. http://dx.doi.org/10.1080/15476286.2016.1191737 (2016).
Poirier, M. C. Antisera specific for carcinogen-DNA adducts and carcinogen-modified DNA: applications for detection of xenobiotics in biological samples. Mutat. Res. 288, 31–38 (1993).
Strickland, P. T. & Boyle, J. M. Immunoassay of carcinogen-modified DNA. Prog. Nucleic Acid. Res. Mol. Biol. 31, 1–58 (1984).
Sawicki, D. L., Erlanger, B. F. & Beiser, S. M. Immunochemical detection of minor bases in nucleic acids. Science 174, 70–72 (1971).
Vold, B. S., Longmire, M. E. & Keith, D. E. Jr. Thiolation and 2-methylthio- modification of Bacillus subtilis transfer ribonucleic acids. J. Bacteriol. 148, 869–876 (1981).
Milstone, D. S., Vold, B. S., Glitz, D. G. & Shutt, N. Antibodies to N6-(Δ2-isopentenyl) adenosine and its nucleotide: interaction with purified tRNAs and with bases, nucleosides and nucleotides of the isopentenyladenosine family. Nucleic Acids Res. 5, 3439–3455 (1978).
Munns, T. W., Liszewski, M. K. & Hahn, B. H. Antibody-nucleic acid complexes. Antigenic domains within nucleosides as defined by solid-phase immunoassay. Biochemistry 23, 2958–2964 (1984).
Horowitz, S., Horowitz, A., Nilsen, T. W., Munns, T. W. & Rottman, F. M. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl Acad. Sci. USA 81, 5667–5671 (1984). This paper uses an antibody to show the presence of m6A in mammalian mRNA.
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012). References 65 and 66 are two pioneering studies combining antibody-based pulldown and high-throughput sequencing.
Mishima, E. et al. Immuno-northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS ONE 10, e0143756 (2015).
Delatte, B. et al. RNA biochemistry. Transcriptome- wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013).
Jacinto, F. V., Ballestar, E. & Esteller, M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 44, 35–43 (2008).
Chen, K. et al. High-resolution N6 -methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. 54, 1587–1590 (2015).
Arluison, V., Buckle, M. & Grosjean, H. Pseudouridine synthetase Pus1 of Saccharomyces cerevisiae: kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA. J. Mol. Biol. 289, 491–502 (1999).
Müller, S. et al. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res. 41, 8615–8627 (2013).
Motorin, Y., Lyko, F. & Helm, M. 5-Methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 38, 1415–1430 (2010).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013).
Metodiev, M. D. et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 10, e1004110 (2014).
Khoddami, V. & Cairns, B. R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013).
Khoddami, V. & Cairns, B. R. Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat. Protoc. 9, 337–361 (2014).
Haag, S. et al. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21, 1532–1543 (2015).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Gu, X., Liu, Y. & Santi, D. V. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl Acad. Sci. USA 96, 14270–14275 (1999).
Spedaliere, C. J. & Mueller, E. G. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 10, 192–199 (2004).
Cahova, H., Winz, M. L., Hofer, K., Nubel, G. & Jaschke, A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011).
Qin, Y. et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22, 111–128 (2016).
Nottingham, R. M. et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22, 597–613 (2016).
Kielpinski, L. J., Boyd, M., Sandelin, A. & Vinther, J. Detection of reverse transcriptase termination sites using cDNA ligation and massive parallel sequencing. Methods Mol. Biol. 1038, 213–231 (2013).
Sooknanan, R., Pease, J. & Doyle, K. Novel methods for rRNA removal and directional, ligation-free RNA-seq library preparation. Nat. Methods 7 http://www.nature.com/app_notes/nmeth/2010/101310/full/nmeth.f.313.html (2010).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 44, e135 (2016).
Birkedal, U. et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 54, 451–455 (2015).
Taoka, M. et al. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. 44, 8951–8961 (2016).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Munafo, D. B. & Robb, G. B. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16, 2537–2552 (2010).
Raabe, C. A. et al. The rocks and shallows of deep RNA sequencing: examples in the Vibrio cholerae RNome. RNA 17, 1357–1366 (2011).
Shen, Y., Zheng, K. X., Duan, D., Jiang, L. & Li, J. Label-free microRNA profiling not biased by 3′ end 2′-O-methylation. Anal. Chem. 84, 6361–6365 (2012).
Mourato, L. L., Beland, F. A. & Marques, M. M. 32P-postlabeling of N-(deoxyguanosin-8-yl)arylamine adducts: a comparative study of labeling efficiencies. Chem. Res. Toxicol. 12, 661–669 (1999).
Sun, W. J. et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 44, D259–D265 (2016).
Bantscheff, M., Schirle, M., Sweetman, G., Rick, J. & Kuster, B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 (2007).
Addepalli, B., Lesner, N. P. & Limbach, P. A. Detection of RNA nucleoside modifications with the uridine-specific ribonuclease MC1 from Momordica charantia. RNA 21, 1746–1756 (2015).
Cao, X. & Limbach, P. A. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Anal. Chem. 87, 8433–8440 (2015).
Wetzel, C. & Limbach, P. A. Mass spectrometry of modified RNAs: recent developments. Analyst 141, 16–23 (2016).
Ross, R., Cao, X., Yu, N. & Limbach, P. A. Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry. Methods 107, 73–78 (2016).
Wetzel, C. & Limbach, P. A. The global identification of tRNA isoacceptors by targeted tandem mass spectrometry. Analyst 138, 6063–6072 (2013).
Kellner, S. et al. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 42, e142 (2014).
Sakaguchi, Y., Miyauchi, K., Kang, B. I. & Suzuki, T. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 560, 19–28 (2015).
Fu, L. et al. Simultaneous quantification of methylated cytidine and adenosine in cellular and tissue RNA by nano-flow liquid chromatography-tandem mass spectrometry coupled with the stable isotope-dilution method. Anal. Chem. 87, 7653–7659 (2015).
Cai, W. M. et al. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol. 560, 29–71 (2015).
Basanta-Sanchez, M., Temple, S., Ansari, S. A., D'Amico, A. & Agris, P. F. Attomole quantification and global profile of RNA modifications: epitranscriptome of human neural stem cells. Nucleic Acids Res. 44, e26 (2016).
Huang, W. et al. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal. Chem. 88, 1378–1384 (2016).
Helm, M., Florentz, C., Chomyn, A. & Attardi, G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res. 27, 756–763 (1999).
Suzuki, T. & Suzuki, T. Chaplet column chromatography: isolation of a large set of individual RNAs in a single step. Methods Enzymol. 425, 231–239 (2007).
Sharma, S., Watzinger, P., Kotter, P. & Entian, K. D. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 41, 5428–5443 (2013).
Buchhaupt, M. et al. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS ONE 9, e89640 (2014).
Bourgeois, G. et al. Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS ONE 10, e0133321 (2015).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA Biol. 7, 237–247 (2010).
Grosjean, H., Droogmans, L., Roovers, M. & Keith, G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 425, 55–101 (2007).
Keith, G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 77, 142–144 (1995).
Stanley, J. & Vassilenko, S. A different approach to RNA sequencing. Nature 274, 87–89 (1978).
Juhling, F. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2009).
Yu, Y. T., Shu, M. D. & Steitz, J. A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3, 324–331 (1997).
Zhao, X. & Yu, Y. T. Detection and quantitation of RNA base modifications. RNA 10, 996–1002 (2004).
Hengesbach, M., Meusburger, M., Lyko, F. & Helm, M. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA 14, 180–187 (2008).
Pyle, A. M., Chu, V. T., Jankowsky, E. & Boudvillain, M. Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol. 317, 140–146 (2000).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Liu, N. & Pan, T. Probing RNA modification status at single-nucleotide resolution in total RNA. Methods Enzymol. 560, 149–159 (2015).
Liu, N. & Pan, T. Probing N6-methyladenosine (m6A) RNA modification in total RNA with SCARLET. Methods Mol. Biol. 1358, 285–292 (2016).
Saikia, M. et al. A systematic, ligation-based approach to study RNA modifications. RNA 12, 2025–2033 (2006).
Dai, Q. et al. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 35, 6322–6329 (2007).
Hiley, S. L. et al. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res. 33, e2 (2005).
Maden, B. E., Corbett, M. E., Heeney, P. A., Pugh, K. & Ajuh, P. M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77, 22–29 (1995).
Aschenbrenner, J. & Marx, A. Direct and site-specific quantification of RNA 2′-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res. 44, 3495–3502 (2016).
Dong, Z. W. et al. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Kellner, S., Kollar, L. B., Ochel, A., Ghate, M. & Helm, M. Structure-function relationship of substituted bromomethylcoumarins in nucleoside specificity of RNA alkylation. PLoS ONE 8, e67945 (2013).
Kellner, S., Seidu-Larry, S., Burhenne, J., Motorin, Y. & Helm, M. A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA. Nucleic Acids Res. 39, 7348–7360 (2011).
Sakurai, M. et al. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 24, 522–534 (2014).
Müller, M. et al. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 43, 10952–10962 (2015).
Vilfan, I. D. et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J. Nanobiotechnol. 11, 8 (2013).
Wescoe, Z. L., Schreiber, J. & Akeson, M. Nanopores discriminate among five C5-cytosine variants in DNA. J. Am. Chem. Soc. 136, 16582–16587 (2014).
Kellner, S. et al. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. (Camb.) 50, 3516–3518 (2014).
Wasserstein, R. L. & Lazar, N. A. The ASA's statement on p-values: context, process, and purpose. Am. Stat. 70, 129–133 (2016).
Rieder, D., Amort, T., Kugler, E., Lusser, A. & Trajanoski, Z. meRanTK: methylated RNA analysis ToolKit. Bioinformatics 32, 782–785 (2016).
Alon, S. et al. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife 4, e05198 (2015).
Hauenschild, R. et al. CoverageAnalyzer (CAn): a tool for inspection of modification signatures in RNA sequencing profiles. Biomolecules 6, 42 (2016).
Durairaj, A. & Limbach, P. A. Mass spectrometry of the fifth nucleoside: a review of the identification of pseudouridine in nucleic acids. Anal. Chim. Acta 623, 117–125 (2008).
Duffy, E. E. et al. Tracking distinct RNA populations using efficient and reversible covalent chemistry. Mol. Cell 59, 858–866 (2015).
Ladner, J. E. & Schweizer, M. P. Effects of dilute HCl on yeast tRNAPhe and E. coli tRNA1fMet. Nucleic Acids Res. 1, 183–192 (1974).
Schleich, H. G., Wintermeyer, W. & Zachau, H. G. Replacement of wybutine by hydrazines and its effect on the active conformation of yeast tRNAPhe. Nucleic Acids Res. 5, 1701–1713 (1978).
Peattie, D. A. Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA 76, 1760–1764 (1979).
Wintermeyer, W. & Zachau, H. G. Tertiary structure interactions of 7-methylguanosine in yeast tRNA Phe as studied by borohydride reduction. FEBS Lett. 58, 306–309 (1975).
Xing, F., Hiley, S. L., Hughes, T. R. & Phizicky, E. M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 279, 17850–17860 (2004).
Plumbridge, J. A., Baumert, H. G., Ehrenberg, M. & Rigler, R. Characterisation of a new, fully active fluorescent derivative of E. coli tRNA Phe. Nucleic Acids Res. 8, 827–843 (1980).
Watson, B. S. et al. Macromolecular arrangement in the aminoacyl-tRNA.elongation factor Tu.GTP ternary complex. A fluorescence energy transfer study. Biochemistry 34, 7904–7912 (1995).
Igloi, G. L. & Kossel, H. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronicacid. Nucleic Acids Res. 13, 6881–6898 (1985).
Igloi, G. L. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27, 3842–3849 (1988).
Acknowledgements
This work was supported by joint Agence Nationale de la Recherche and Deutsche Forschungsgemeinschaft (ANR-DFG) grants for High-throughput technology for detection of RNA modifications (HTRNAMod) (ANR-13-ISV8-0001/HE 3397/8-1) and by the DFG priority programme SPP1784.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- RNA modifications
-
Chemically altered (modified) nucleotides in mature RNA chain.
- Nucleotides
-
The smallest structural units of a nucleic acid, composed of a nucleobase connected to a ribose (or deoxyribose) via a glycosidic bond, and a phosphate esterified to a hydroxyl group of the sugar moiety. The combination of only nucleobase and sugar is referred to as a nucleoside.
- Reverse transcription
-
The synthesis of single-stranded DNA (known as complementary DNA (cDNA)) by reverse transcriptase (an RNA-dependent DNA polymerase) using single-stranded RNA as a template.
- RT signature
-
A specific pattern composed of misincorporation in cDNA and reverse transcription arrest of primer extension, induced by modified residues in an RNA chain.
- Haptens
-
Small, otherwise immunosilent molecules that elicit an immune response only when exposed to the immune system in the form of a covalent conjugate to a large carrier such as a protein.
- Pulldown
-
A technique for selective isolation of RNA (or protein) species from a complex biological sample. It generally relies on affinity capture with antibodies or another high-affinity interaction.
- Click chemistry
-
A class of biocompatible (bio-orthogonal) reactions, generally involving azide and alkyne moieties.
- Barcodes
-
Short specific sequences (often six to eight nucleotides) that are incorporated into sequencing adaptors and ligated to samples. When barcode sequences are shared within a sample but differ between samples, they allow unambiguous retrospective assignment of samples run in the same sequencing lane. When barcodes are degenerate and highly variable within a sample, they are known as unique molecular identifiers (UMIs) and are used to identify redundant, PCR-amplified reads that originate from the same reverse transcription event. Adaptors may combinatorially contain both sample-specific barcodes and UMIs.
- CircLigation
-
Circularization of single-stranded cDNA templates having a 5′ phosphate and a 3′ hydroxyl group with CircLigase.
- Receiver operating characteristics curve
-
(ROC curve). A graphical plot illustrating the performance of a binary classifier system as a function of varied thresholds for true positives and false positives. Plotting the true-positive rate (TPR) against the false-positive rate (FPR) at various threshold settings leads to the ROC curve, in which the area under the curve (AUC) is a measure of the performance. The true-positive rate is also known as sensitivity, the FPR can be calculated as 1 – specificity.
- Lipophilicity
-
'Fat-loving' character of a compound, reflected in its ability to dissolve in nonpolar solvents.
- DNAzymes
-
Artificially generated DNA sequences that contain catalytic activity, for example, RNase activity.
- Thin-layer chromatography
-
(TLC). A technique for separation carried out in a thin layer of adsorbent material, also known as a stationary phase. TLC can be one-dimensional (1D-TLC) if it involves one type of separation or two-dimensional (2D-TLC) if it involves separation in one dimension using one solvent, followed by separation in a second dimension using a second solvent that separates on the basis of distinct physicochemical properties.
- Splinted ligation
-
A technique to biochemically join nucleic acid fragments by enzymatic ligation. Both fragments are brought in spatial proximity by hybridization to a so-called splint DNA, which is an oligodeoxynucleotide that is essentially a cDNA of the target RNA molecule.
- Nanopore technology
-
Technology that funnels molecules, including nucleic acids, through nanosized pores (typically protein-based pores), thereby opening the possibility of determining their sequence (and potentially also a subset of their covalent modifications).
Rights and permissions
About this article
Cite this article
Helm, M., Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 18, 275–291 (2017). https://doi.org/10.1038/nrg.2016.169
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2016.169
This article is cited by
-
Clinical significance of RNA methylation in hepatocellular carcinoma
Cell Communication and Signaling (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Spatial imaging of glycoRNA in single cells with ARPLA
Nature Biotechnology (2024)
-
RNA modification: mechanisms and therapeutic targets
Molecular Biomedicine (2023)
-
Navigating the pitfalls of mapping DNA and RNA modifications
Nature Reviews Genetics (2023)