Abstract
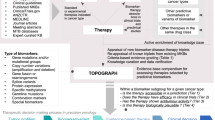
As diagnostic tests become increasingly important for optimizing the use of drugs to treat cancers, the co-development of a targeted therapy and its companion diagnostic test is becoming more prevalent and necessary. In July 2011, the US Food and Drug Administration released a draft guidance that gave the agency's formal definition of companion diagnostics and introduced a drug–diagnostic co-development process for gaining regulatory approval. Here, we identify areas of drug–diagnostic co-development that were either not covered by the guidance or that would benefit from increased granularity, including how to determine when clinical studies should be limited to biomarker-positive patients, defining the diagnostically selected patient population in which to use a companion diagnostic, and defining and clinically validating a biomarker signature for assays that use more than one biomarker. We propose potential approaches that sponsors could use to deal with these challenges and provide strategies to help guide the future co-development of drugs and diagnostics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Naylor, S. & Cole, T. Overview of companion diagnostics in the pharmaceutical industry. Drug Discovery World [online], (2010).
US National Cancer Institute. Targeted cancer therapies (Fact Sheet). US National Cancer Institute [online], (2012).
Savage, D. G. & Antman, K. H. Imatinib mesylate — a new oral targeted therapy. N. Engl. J. Med. 346, 683–693 (2002).
Demetri, G. D. et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 347, 472–480 (2002).
Garnock-Jones, K. P., Keating, G. M. & Scott, L. J. Trastuzumab: a review of its use as adjuvant treatment in human epidermal growth factor receptor 2 (HER2)-positive early breast cancer. Drugs 70, 215–239 (2010).
Hudziak, R. M. et al. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol. Cell. Biol. 9, 1165–1172 (1989).
Kulke, M. H. Systemic therapy for advanced pancreatic neuroendocrine tumors. Semin. Oncol. 40, 75–83 (2013).
Motzer, R. J. et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA 295, 2516–2524 (2006).
US Food & Drug Administration. Guidance for industry and Food and Drug Administration staff — in vitro companion diagnostic devices (draft guidance). US Food & Drug Administration [online], (2011).
Teutsch, S. M. et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet. Med. 11, 3–14 (2009).
US Food & Drug Administration. Guidance for industry: enrichment strategies for clinical trials to support approval of human drugs and biological products (draft duidance). US Food & Drug Administration [online], (2012).
European Medicines Agency. Reflection paper on co-development of pharmacogenomic biomarkers and assays in the context of drug development. European Medicines Agency [online], (2010).
US Food & Drug Administration. Drug-diagnostic co-development concept paper (draft — not for implementation). US Food & Drug Administration [online], (2005).
Moore, M. W., Babu, D. & Cotter, P. D. Challenges in the codevelopment of companion diagnostics. Per. Med. 9, 485–496 (2012).
Beckman, R. A., Clark, J. & Chen, C. Integrating predictive biomarkers and classifiers into oncology clinical development programmes. Nature Rev. Drug Discov. 10, 735–748 (2011).
Ravnan, M. C. & Matalka, M. S. Vemurafenib in patients with BRAF V600E mutation-positive advanced melanoma. Clin. Ther. 34, 1474–1486 (2012).
Tanizaki, J. et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin. Cancer Res. 18, 6219–6226 (2012).
Brand, T. M., Iida, M. & Wheeler, D. L. Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biol. Ther. 11, 777–792 (2011).
Chung, K. Y. et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J. Clin. Oncol. 23, 1803–1810 (2005).
Fridlyand, J. et al. An industry statistician's perspective on PHC drug development. Contemp. Clin. Trials http://dx.doi.org/10.1016/j.cct.2013.04.006 (2013).
Freidlin, B., McShane, L. M., Polley, M. Y. & Korn, E. L. Randomized phase II trial designs with biomarkers. J. Clin. Oncol. 30, 3304–3309 (2012).
Kaiser, L., Becker, C., Kukreti, S. & Fine, B. Decision making for a companion diagnostic in an oncology clinical development program. Drug Inform. J. 46, 294–302 (2012).
Mandrekar, S. J. & Sargent, D. J. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J. Clin. Oncol. 27, 4027–4034 (2009).
Mandrekar, S. J. & Sargent, D. J. All-comers versus enrichment design strategy in phase II trials. J. Thorac. Oncol. 6, 658–660 (2011).
Simon, R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Per. Med. 7, 33–47 (2010).
Zhan, S. et al. Defective neuropeptide processing and ischemic brain injury: a study on proprotein convertase 2 and its substrate neuropeptide in ischemic brains. J. Cereb. Blood Flow Metab. 29, 698–706 (2009).
Spiegl-Kreinecker, S. et al. O6-methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol. 12, 28–36 (2010).
Brannath, W. et al. Confirmatory adaptive designs with Bayesian decision tools for a targeted therapy in oncology. Stat. Med. 28, 1445–1463 (2009).
Chen, C. & Beckman, R. A. Hypothesis testing in a confirmatory phase III trial with a possible subset effect. Stat. Biopharm. Res. 1, 431–440 (2009).
Hochberg, Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75, 800–802 (1988).
Wang, S. J., O'Neill, R. T. & Hung, H. M. Approaches to evaluation of treatment effect in randomized clinical trials with genomic subset. Pharm. Stat. 6, 227–244 (2007).
Jiang, W., Freidlin, B. & Simon, R. Biomarker-adaptive threshold design: a procedure for evaluating treatment with possible biomarker-defined subset effect. J. Natl Cancer Inst. 99, 1036–1043 (2007).
Christine, M. M., Sharly, J. N. & Gilbert, S. O. (eds) Evolution of Translational Omics: Lessons Learned and the Path Forward (The National Academies Press, 2012).
Breiman, L. Bagging predictors. Machine Learn. 24, 123–140 (1996).
Freidlin, B., Jiang, W. & Simon, R. The cross-validated adaptive signature design. Clin. Cancer Res. 16, 691–698 (2010).
Freidlin, B. & Simon, R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin. Cancer Res. 11, 7872–7878 (2005).
Kaiser, L. D. Stratification of randomization is not required for a pre-specified subgroup analysis. Pharm. Stat. 12, 43–47 (2013).
Scher, H. I., Nasso, S. F., Rubin, E. H. & Simon, R. Adaptive clinical trial designs for simultaneous testing of matched diagnostics and therapeutics. Clin. Cancer Res. 17, 6634–6640 (2011).
Simon, R. M., Paik, S. & Hayes, D. F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 101, 1446–1452 (2009).
European Medicines Agency. Reflection paper on methodological issues associated with pharmacogenomic biomarkers in relation to clinical development and patient selection. European Medicines Agency [online], (2011).
Goldberg, P. KRAS finding changes oncology practice but poses profound regulatory dilemma. The Cancer Letter 35, 1–8 (2009).
Patterson, S. D. et al. Prospective-retrospective biomarker analysis for regulatory consideration: white paper from the industry pharmacogenomics working group. Pharmacogenomics 12, 939–951 (2011).
Simon, R. & Roychowdhury, S. Implementing personalized cancer genomics in clinical trials. Nature Rev. Drug Discov. 12, 358–369 (2013).
Park, R. Companion diagnostics: a blossoming field. IVD Technology [online], (2011).
Gaffney, A. Device group launches offensive against pending EU legislation. Regulatory Focus [online], (2013).
European Commission. In vitro diagnostic medical devices: Directive 98/79/EC. European Commission [online], (1998).
Stynen, D. Revision of Europe's IVD Directive 98/79/EC. IVD Technology [online], (2011).
European Commission. Revision of the medical device directives. European Commission [online], (2012).
Study Group 1 of the Global Harmonization Task Force. Principles of In Vitro Diagnostic (IVD) Medical Devices Classification [online], (Global Harmonization Task Force, 2008).
Acknowledgements
The authors would like to thank the following for their contributions to, and critical review of, this manuscript: S. Averbush, R. Beckman, L. Burdette, T. Bush, R. Canetta, S. Dahm, J. Dudinak, L. Farrington, S. Ford, G. Hampton, L. Hashimoto, D. Hayes, S. Ho, F. Houn, E. Ibia, J. Jenkins-Showalter, L. Lavange, G. Lieberman, M. Liu, S. Lutzker, P. Mahaffy, L. Mansfield, A.-M. Martin, I. McCaffery, D. Miller, V. Miller, A. Mueller, P. Paoletti, S. Patterson, C. Paulding, R. Pazdur, D. Rasmussen, J. Roche, S. Scherer, J. Shuren, J. Siegel, G. Spaniolo, B. Trepicchio, W. Verbiest, J. Wiezorek, J. Woodcock and L. Zydowski.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.F. is employed by Genentech.
R.B. is employed by Pfizer.
D.P.S. is employed by Agios Pharmaceuticals.
All other authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Threshold readjustment using a split-dataset approach (PDF 111 kb)
Glossary
- Bootstrap aggregating
-
A type of model averaging that improves the stability and accuracy of the algorithms that are used in biomarker studies, and is typically conducted by repeatedly re-sampling data points from a given data set.
- Bridging studies
-
Studies in which clinical samples that were tested initially with an assay used in a clinical trial are re-tested with another assay to support the approval or clearance of that assay.
- Classifier
-
An algorithm (or statistical rule) that can be used to predict prognosis or the responsiveness of patients to a given therapy and thereby used to select and/or stratify patients for therapy in clinical trials. The inputs to the algorithm are the values obtained from one or more predefined biomarkers.
- Diagnostic platform
-
A form of molecular diagnostic testing that provides patient-specific information using parallelized platform sequencing technology.
- Effect size
-
An estimate of the treatment effect relative to the control (or any other parameter of interest).
- Hierarchical approaches
-
Sequential approaches to the testing of multiple hypotheses where a given null hypothesis can only be tested if all null hypotheses that are ranked higher are rejected.
- Investigational device exemption
-
A regulatory submission that allows a medical device to be used in a clinical study without full approval from the US Food and Drug Administration in order to collect data on the safety and effectiveness of the device.
- Laboratory-developed tests
-
A class of in vitro diagnostic tests that are currently not regulated by the US Food and Drug Administration.
- Next-generation sequencing
-
A set of technologies that enable the rapid generation of enormous amounts of DNA or RNA sequencing data.
- Notified body
-
An organization that has been accredited by a member country of the European Union to determine whether a product meets certain predetermined standards.
- Split-α approaches
-
Approaches that are undertaken for the testing of multiple hypotheses to maintain the study-wise type I error at the intended 0.05 level by splitting the threshold for declaring significance (that is, α) among the hypotheses to be tested.
- Statistical analysis plan
-
The pre-specified analyses that will be applied to the data generated from a clinical trial.
- Summary measures
-
The mathematical combination of values produced by one or more biomarkers, resulting in a single value that can be used for making decisions about drug treatments.
- Training set
-
A data set that is used for the development of a statistical model and all of its parameters. Another data set known as the test set is then used to test the accuracy of the model.
- Type I error
-
The chance of falsely rejecting the null hypothesis in favour of the alternative (the probability of the false positive); for example, falsely claiming that a relationship exists between treatment effect and biomarker value in the absence of a true relationship.
- Unbiased effect estimate
-
An estimate of the treatment effect in which the expected value (based on hypothetical repetitions of the study) equals the true value of the effect.
Rights and permissions
About this article
Cite this article
Fridlyand, J., Simon, R., Walrath, J. et al. Considerations for the successful co-development of targeted cancer therapies and companion diagnostics. Nat Rev Drug Discov 12, 743–755 (2013). https://doi.org/10.1038/nrd4101
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4101
This article is cited by
-
Changes in Companion Diagnostic Labelling: Implementation of FDA’s April 2020 Guidance for Industry for In Vitro CDx Labeling for Specific Oncology Therapeutic Groups
Therapeutic Innovation & Regulatory Science (2022)
-
How are we evaluating the cost-effectiveness of companion biomarkers for targeted cancer therapies? A systematic review
BMC Cancer (2021)
-
Annotation and detection of drug effects in text for pharmacovigilance
Journal of Cheminformatics (2018)
-
Integrative omics analyses broaden treatment targets in human cancer
Genome Medicine (2018)
-
Molecular classification of tissue from a transformed non-Hogkin’s lymphoma case with unexpected long-time remission
Experimental Hematology & Oncology (2017)