Key Points
-
Bacteria use many virulence factors to cause disease in the human host, including adhesins that bind to host cells and facilitate colonization, the production of toxins that kill or change signal transduction in mammalian cells, and the production of specialized secretion systems to deliver effectors (bacterial proteins that often function as molecular mimics of eukaryotic proteins).
-
Expression of these metabolically expensive virulence traits is precisely regulated. In many cases this regulation exploits chemical signalling between bacterial cells, and bacteria and the host (inter-kingdom chemical signalling).
-
Anti-virulence therapeutic strategies interfere with these virulence traits, preventing bacterial pathogenesis without killing the bacteria.
-
Anti-virulence approaches may engender a milder evolutionary pressure towards the development of resistance than conventional antibiotics.
-
Anti-virulence strategies have been developed to prevent bacterial adhesion to host tissue, interfere with bacterial toxins and modulate virulence gene expression and bacterial chemical communication.
-
Many of these approaches explore novel drug targets.
-
Although promising, most of these anti-virulence drugs are still at the basic research level, and clinical trials have yet to be conducted.
Abstract
Antibiotic resistance is one of the greatest challenges of the twenty-first century. However, the increasing understanding of bacterial pathogenesis and intercellular communication has revealed many potential strategies to develop novel drugs to treat bacteria-mediated disease. Interference with bacterial virulence and/or cell-to-cell signalling pathways is an especially compelling approach, as it is thought to apply less selective pressure for the development of bacterial resistance than traditional strategies, which are aimed at killing bacteria or preventing their growth. Here, we discuss the mechanisms of bacterial virulence and present promising anti-virulence strategies and compounds for the future treatment of bacterial infections.
Similar content being viewed by others
Main
Infectious diseases are the leading cause of death worldwide, with bacterial infections contributing substantially to this high rate of mortality. Although in 1969 the General Surgeon of the United States proclaimed that “We have closed the chapter on infectious diseases, due to antibiotics”, many bacteria are now multidrug resistant.
There are a wide range of factors that the invading bacteria must overcome to cause disease. These include innate factors such as pH, temperature and host secretions, as well as active factors such as the immune response, which act to prevent bacterial colonization. In addition to the host factors are the existing bacterial microbiota that are resident at most anatomical niches in which infection occurs. This resident microbiota must be removed or exploited for a successful infection. Bacterial pathogens use many diverse strategies to cause disease in human hosts1.
Traditional approaches to combat bacterial infection were primarily identified in and derived from other microorganisms. These approaches rely on the disruption of the growth cycle by preventing the synthesis and assembly of key components of bacterial processes such as cell wall synthesis, DNA replication and protein synthesis2. Although these strategies and compounds were highly effective, they resulted in substantial stress on the target bacterium, which rapidly selected for resistant subpopulations3. For example, in a population of ∼10 billion bacteria, a minute portion will be resistant to a given antibiotic, owing to natural bacterial mutation rates. If the majority of the non-resistant cells are killed, the smaller fraction that is resistant may quickly grow and become the dominant proportion of the population.
Consequently, we are now entering the post-antibiotic era, with limited treatment options for many bacterial infections, such as the now widely disseminated vancomycin-resistant Enterococcus species3 and methicillin-resistant Staphylococcus aureus4 (MRSA). Although these two pathogens are by no means the only examples, they are the most recognizable in the popular literature and prove that inappropriate and indiscriminate antibiotic treatment can lead to resistant organisms in a short period of time. However, many alternative strategies are currently being investigated that are based on the inhibition of virulence rather than of bacterial growth. Such strategies might apply a milder evolutionary pressure for the development of resistance as most virulence traits are not essential for bacterial survival.
Bacteria sense their environment and, once in the host, they respond by initiating a defined programme to activate virulence traits5,6. Anti-virulence signalling strategies may specifically interfere with the ability of the bacteria to recognize host signals that alert the bacteria that they are at the site of infection and/or activate specific virulence traits that are needed to establish infection. By preventing the expression or activity of virulence traits, the bacteria are less able to colonize the host. In addition, as this strategy does not directly kill the bacteria, there is presumably less evolutionary pressure for the development of resistant clones than with traditional antibiotics. It is thought that this inhibition could allow the host immune system, including the normal microbiota, to prevent bacterial colonization or clear any established infection. In addition, these anti-virulence drugs could potentially be used in combination with established or novel antimicrobials in a synergistic manner to extend the lifespan of these drugs7,8. One recent promising strategy relies on newly discovered synthetic or natural small organic compounds that serve as a generation of novel anti-virulence drugs5,9.
This Review provides a brief overview of the mechanisms that bacteria use to cause disease in a host. Promising anti-virulence strategies to overcome these mechanisms that are currently under investigation and possible future approaches are discussed.
Molecular mechanisms of bacterial virulence
Bacteria use an array of virulence factors to cause disease in the human host. These factors include adhesins, which bind to host cells and facilitate colonization; toxins, which kill or change signal transduction in mammalian cells; and specialized secretion systems to deliver effectors. Because expression of these virulence traits is metabolically expensive, gene expression is exquisitely regulated so that it only occurs at the right site for colonization in the host (Table 1). Here, we introduce and review these mechanisms of bacterial virulence.
Toxins. Broad arrays of toxins are secreted by bacterial species, resulting in the alteration of eukaryotic host cells and the development of disease. These toxins are often identified as the first virulence factors in pathogenesis, and have been studied in depth. Examples of such toxins include the pertussis toxin from Bordetella pertussis, which is the cause of whooping cough (recently reviewed in Ref. 10); toxins of Clostridium difficile, which cause diarrhoea11; and many others that are discussed in this Review. We have selected two toxin systems that are well known and have received considerable attention in the production of anti-toxin therapeutics. The classical toxins, such as Shiga toxin from Escherichia coli12 and anthrax toxin from Bacillus anthracis, are known as AB toxins because they are composed of the active subunit (A subunit) and a multimeric receptor-binding subunit (B subunit)13 (Fig. 1). The B subunit adheres to a surface-exposed receptor on the eukaryotic cell, and the A subunit enters the cell, in which it exerts its enzymatic activity. The Shiga toxin B subunit binds to the globosylceramide 3 (Gb3) ganglioside that is abundant on kidney and brain endothelial cells, whereas the A subunit inhibits host protein synthesis by cleaving a glycosidic bond on the 28S ribosomal RNA in the 60S subunit of the ribosome14. By contrast, anthrax toxin is a tripartite toxin that has a single binding subunit known as the protective antigen (PA) and two active subunits known as the lethal factor (LF) and the edema factor (EF). The PA subunit is secreted from the bacterium as an 83 kDa subunit, which binds to the host cell surface. It is cleaved by a host protease to a 63 kDa subunit that can assemble into the heptameric ring in the lysosome wall, in which it forms a pore enabling the extrusion of LF and EF into the host cell cytoplasm15. Once secreted into the cytoplasm, the LF functions as a metalloprotease that cleaves mitogen-activated protein kinase kinases (MAPKKs), resulting in altered signalling in the host cell. EF is an adenylyl cyclase enzyme that, when activated by calmodulin, dramatically increases the cyclic AMP levels in the cell. The net effect of EF in human tissue is the accumulation of fluid in extracellular spaces16. These are only two examples of toxin activities; however, each has a number of steps that could be therapeutically inhibited (Fig. 1).
The major steps in the secretion of a bacterial AB toxin such as anthrax toxin or Shiga toxin are shown. Strategies for therapeutic intervention are illustrated in grey boxes. a | Anthrax toxin is a tripartite toxin composed of the protective antigen (PA), the edema factor (EF) and the lethal factor (LF). PA binds to the host receptors anthrax toxin receptor 1 (ANTXR1; also known as ATR and TEM8) and ANTXR2 (also known as CMG2), and is then cleaved by host factors. The cleaved PA can form a heptameric complex, which interacts with either EF or LF. Once the completed toxin is formed, it can be actively or passively taken up into the eukaryotic cell. The active subunits interact with specific target cell components and cause a response in the host cell. EF is activated upon binding to the host cell calmodulin and then acts as an adenylate cyclase, increasing the intracellular levels of cyclic AMP, which leads to changes in signal transduction and fluid accumulation. LF is a metalloprotease that cleaves mitogen-activated protein kinase kinase (MAPKK), leading to cell death. Many inhibitors of anthrax toxin have been developed: soluble PA analogues and PA-specific antibodies inhibit PA binding to the host receptors; small-molecule inhibitors block the formation of the pore in the host cell membrane by the heptameric PA complex; small molecules inhibit EF and LF entry into the host cell; and small molecules and cisplatin prevent LF function. b | Shiga toxin is composed of an A subunit, which is enzymatically active, and a B subunit, which forms a pentamer that binds to the host receptor globosylceramide 3 (Gb3). The A subunit cleaves a glycosidic bond on the 28S ribosomal RNA, stopping protein synthesis and leading to cell death. Shiga toxin causes haemolytic uraemic syndrome (HUS). The Shiga toxin inhibitor Synsorb-Pk mimics the Gb3 receptor. Binding of Synsorb-Pk to the B subunit of Shiga toxin prevents the toxin binding to the host receptor.
Adhesins and biofilms. To effectively colonize the host and promote disease, bacteria must attach to host cells. Most bacteria have a specific host range and will only infect hosts that express particular receptors for bacterial adhesion factors on their cell surface. Furthermore, once within the host, bacteria will infect only the subset of cells or tissues that express the appropriate receptor. The human body has a number of innate responses, such as peristaltic motion in the gastrointestinal tract and the coordinated beating of cilia in the nasopharyngeal regions, which remove bacteria that do not have the appropriate adhesive capabilities17. The attachment of bacteria to a host cell is an intricate process governed by the adhesin on the bacteria and the receptor on the host cell18 (Fig. 2). Adherence is often only the first step in the infection process, which also involves internalization, deeper tissue penetration and possible systemic distribution18. Bacteria use many types of appendages for adhering to the host surface including, but not limited to, pili, fimbriae and in some cases flagella18. In many cases, the binding structure is a pilus — a long multisubunit appendage that often has binding specificity in the terminal subunits. Although many pili have been identified for several pathogens, the most studied adhesins are encoded by uropathogenic E. coli (UPEC), which is the leading cause of urinary tract infections19. UPEC encodes the type I pili19 and the P pili (encoded by the pap pilus operon), both of which are required during infection for adhesion to the urogenital epithelia, to prevent the bacterium from being washed out with the normal flow of urine20. The gene cluster encoding the P pilus is often found in multiple copies in particularly virulent isolates of UPEC21.
Bacteria adhere to host cells to colonize the host and initiate infection. Therapeutic inhibitors are shown as red circles and strategies to block bacteria are described. a | Some of the most common adhesins are pili or fimbriae, which are 'hair-like' structures that adhere to specific host receptors. Adhesion can be inhibited by preventing the assembly of the adhesion complex, as is the case for the P pili of uropathogenic Escherichia coli. b | A schematic depiction of pili assembly in the surface of bacteria. The pili subunits polymerize to form a pilus. Inhibitors of biogenesis are mimics of the normal pilin subunits that, when incorporated into the growing pilus, prevent elongation and the formation of a functional pilus.
Bacteria also form biofilms, which are communities of bacteria encased within hydrated polymeric matrices. Biofilms are used to adhere to living or inert surfaces and provide protection from the immune system. Furthermore, they are refractory to antimicrobial compounds22. Biofilms have been linked to a growing list of medically relevant pathogens including Pseudomonas aeruginosa, which causes several types of nosocomial infection and cystic fibrosis23, and S. aureus, which causes a wide range of diseases from minor skin infections to toxic shock syndrome24.
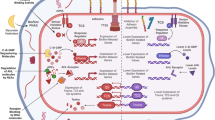
Specialized secretion systems. Many bacterial pathogens produce a specialized secretory system that resembles a syringe and is used by bacteria to inject toxins (effectors) into the host cell (Fig. 3). These effectors mimic the function of host proteins and substantially alter mammalian signalling pathways, which contributes to the disease process25. Three distinct secretory systems, type III, IV and VI, are involved in the translocation of bacterial effectors directly into the host cells26. The type III secretion system (TTSS) consists of several proteins forming a needle-type structure that permits bacteria to inject effectors directly from the bacterial cytoplasm into the eukaryotic cell cytoplasm. These secreted effectors often alter the signal transduction in the host cell to enhance microbial survival, invasion or attachment27,28. They can cause inflammation, opening of tight junctions (for example, by enteropathogenic E. coli), electrolyte secretion and alterations in the cytoskeleton (for bacterial invasion by, for example, Shigella and Salmonella species and for bacterial adhesion of, for example, enteropathogenic E. coli)26. By contrast, the type IV secretory system is used to transfer either bacterial DNA (for example, by Agrobacteria tumefaciens, which is a plant pathogen) or bacterial effector proteins to mammalian cells (for example, by Helicobacter pylori). This system also forms a channel between the bacterial and eukaryotic cell cytoplasm. It is a long pilus-like structure rather than a needle structure26. Type VI secretion systems form tubular structures, but precisely how these systems assemble and deliver effectors into the host cell remains largely unknown29.
Prokaryotic cells use active secretion to inject effector molecules into the eukaryotic cell. Three systems are currently known to be capable of this: type III, IV and VI. Each of these systems has minor differences in structure and gene content but, through the liberation of energy by the hydrolysis of ATP, they all deliver the effectors directly into the eukaryotic cytoplasm. Type III secretion systems resemble syringes that inject effectors into the host cell, type IV secretion systems use a pilus-like structure to inject effectors into host cells, whereas type VI secretion systems are tubular structures and are the most recently described. Precisely how the type VI system assembles and delivers effectors into the host cell remains largely unknown. Once within the host cell, these effectors change multiple signal transduction pathways that affect the actin cytoskeleton, cause inflammation and change electrolyte influx into the cell. Strategies to therapeutically inhibit these bacterial secretion systems are shown in grey boxes.
Regulation of virulence gene expression. Bacteria tightly regulate expression of metabolically expensive virulence traits to avoid wasting energy. Regulation of the expression of virulence factors can occur at many levels and can share similarities among a wide range of bacterial species30. Most of the underlying mechanisms for this regulation require sensing of a specific signal in the environment6. The specific mechanisms are too numerous to be completely discussed in this Review. A growing number and range of bacterial signalling molecules has been identified over recent years as scientists have become more sophisticated in identifying them and recognizing their sometimes subtle effects31. These signals can be environmental cues, such as temperature and osmolarity, or chemicals produced by other bacteria in the host. However, in many cases, the signals to which bacteria respond are provided by the potential host species in a type of cell-to-cell communication known as inter-kingdom signalling6,32. Some of these signalling pathways are conserved in many bacterial species, and targeting of these systems for therapeutics may yield broad-spectrum drugs. The various chemical cell-to-cell signalling mechanisms that are used by bacteria are collectively known as quorum-sensing systems33. The first quorum-sensing system was identified in Vibrio fisheri, a bacterium that controls bioluminescence production in response to small molecules that gauge bacterial population cell density34,35. P. aeruginosa is a bacterial pathogen that causes opportunistic infections in burns, wounds, ventilator-associated pneumonia and in patients with cystic fibrosis. P. aeruginosa encodes a wide range of virulence factors including a TTSS, proteases, adhesins and the ability to form biofilms. These virulence traits are hierarchically and precisely regulated by quorum sensing36. P. aeruginosa has several quorum-sensing mechanisms, which depend on the release of chemical signals that are divided into two groups based on their chemistry. The first group, the acyl homoserine lactones (AHLs) includes N-3-oxo-dodecanoyl-L-homoserine lactone (C12-HSL) and N-butanoyl-L-homoserine lactone (C4-HSL). The second group, the 4-quinolones, forms the Pseudomonas quinolone signal (PQS)37,38,39,40,41. These signals are produced and sensed by three quorum-sensing systems, LasR–LasI and RhlR–RhlI for the AHLs, and PqsR–pQSABCDE for the PQS signal37,39,40,42,43. LasI and RhlI synthesize C12-HSL and C4-HSL, respectively, whereas pQSABCD synthesizes the PQS signal40,42. These signals bind to transcription regulators (C12-HSL binds to the LasR receptor, C4-HSL binds to the RhlR receptor and PQS binds to the PqsR LysR-like receptor37,39,40,42,43) and induce the expression of virulence genes and biofilm formation (Fig. 4A).
The mechanisms of action of inhibitors that specifically target virulence signalling are shown. a | Inhibitors of acyl homoserine lactone (AHL) and Pseudomonas quinolone signal (PQS) bacterial cytoplasmic receptors (LasR, RhlR and PqsR) in Pseudomonas aeruginosa. The inhibitors are thought to bind to these receptors and prevent recognition of the bacterial signal that would activate virulence gene expression. b | Inhibitors of the Agr system in Staphylococcus aureus. Autoinducing peptide (AIP) mimics bind to AgrC, preventing recognition of the S. aureus AIP. This inhibits AgrC autophosphorylation and the subsequent transfer of this phosphate group to the AgrB transcription factor, which would otherwise activate a multitude of virulence genes. Therefore, blocking AgrC by AIP inhibitors will prevent activation of virulence signalling. c | QseC inhibitors. QseC senses the bacterial autoinducer 3 (AI-3) signal and the host adrenaline (ADR) and noradrenaline (NA) hormones. Upon sensing these signals, QseC increases its autophosphorylation and transfers this phosphate group to three response regulators (QseB, QseF and KdpE) that activate a complex virulence gene regulation cascade in enterohaemorrhagic Escherichia coli. LED209 binds to QseC, preventing it from binding to its cognate signals to activate the expression of virulence genes.
Therefore, every bacterial genome contains two-component regulatory systems composed of a histidine sensor kinase, which is usually in the membrane of the bacteria, and a response regulator, which is usually a transcription factor activated upon phosphorylation. Histidine kinases are the major environmental sensory proteins in bacteria. Although the ligands for these systems are not well known in most cases, it is thought that these signalling mechanisms can be exploited as conserved therapeutic targets44. Drugs targeting bacterial signalling systems are attractive owing to their selective toxicity, given that these systems are not usually found in the eukaryotic hosts (Figs 4,5). A recent study revealed a common signalling pathway that begins with the membrane-bound QseC histidine sensor kinase5. QseC homologues are present in at least 25 important human and plant pathogens5, and qseC mutants of enterohaemorrhagic E. coli (EHEC)5,45, Salmonella typhimurium5,46 and Francisella tularensis47 have been shown to have decreased virulence in animal models of infection. QseC binds to and increases its autophosphorylation in response to the bacterial quorum-sensing signal autoinducer 3 (AI-3) and/or the host signals adrenaline and/or noradrenaline45. This initiates a complex phosphorylation cascade in the bacterial cell that regulates the expression of virulence genes5,45,48.
a | Inhibitors that specifically target virulence will prevent the transcription and translation of virulence factors. However, other inhibitors could include molecules that inhibit the activity of a virulence factor or prevent the substrate from binding to the appropriate ligand. b | Global inhibitors have activity at upstream stages in a signalling cascade and therefore exert broader changes on the expression pattern of the bacterium than more specific inhibitors. Signalling through the membrane component may be therapeutically inhibited by preventing signal interaction with the receptor, which prevents phosphotransfer, the activation of the response regulator (RR) and any downstream effects. A primary example is the use of LED209 to inhibit global virulence in enterohaemorrhagic Escherichia coli, Salmonella spp. and Francisella tularensis. These mechanisms represent general points of possible inhibition. IM, inner membrane; OM, outer membrane.
Anti-virulence targets and approaches
A novel approach to developing therapies for infectious diseases is to block bacterial virulence specifically, without killing or inhibiting bacterial growth. Many anti-virulence strategies are being explored, including inhibiting bacterial adhesion to the host cell, inhibiting toxins or specialized secretion systems, and interfering with gene regulation of virulence traits (Fig. 6; Table 2). These strategies are discussed below.
For information on the mode of action, development stage and indication of the compounds, see Table 2.
Targeting toxins. Although there are many bacterial toxins that could be discussed, this section will focus on toxins for which an inhibitor has been identified at least in vitro and in many cases in vivo.
Anthrax toxin is a crucial component of B. anthracis infection49, as evidenced by the fact that the attenuated vaccine, known as the Pasteur strain, lacks the ability to produce anthrax toxin49. As a complete lack of the toxin renders the isolate attenuated in humans and large mammals, it follows that inactivation of the toxin should have a similar effect. Antimicrobial therapies have been developed based on the inactivation of the toxin at different stages of processing and/or activation. Two receptors have been identified for the PA subunit on human cells: anthrax toxin receptor 1 (ANTXR1; also known as ATR and TEM8) and ANTXR2 (also known as CMG2)50,51. The simplest strategy to prevent the deleterious effects of the toxin is to block toxin binding to its cognate receptor by the introduction of soluble analogues to the receptor. This has been shown to be effective in the inbred F344 rat model52, but has not yet been attempted in humans. An alternative strategy is the development of antibodies that bind directly to the PA subunit, thereby preventing toxin interaction with the host cell membrane53. Four single-chain variable fragments of antibodies that target the PA subunit of anthrax toxin have been identified using a phage display library and were shown to be protective in the F344 rat model and in chimpanzees53. The antibody subunits could be injected and used for short-term prophylaxis against anthrax toxin in emergency situations.
Compounds designed to block the PA heptamer pore have also been developed, including low-molecular-mass compounds that prevented LF and EF entry into cells54. The synthetic small cyclic molecule per-6-(3-aminopropylthio)-β-cyclodextrin binds to the PA pore lumen, blocking PA-induced transport at subnanomolar concentrations. This compound protected macrophages from the cytotoxicity of anthrax lethal toxin54.
In addition to strategies to block the adhesion and the pore of the PA subunit, considerable effort has focused on identifying inhibitors of the LF subunit. Using a small-molecule inhibitor screen, it was shown that a small-molecule inhibitor, the hydroxamate (2R)-2-[(4-fluoro-3-methylphenyl)sulphonylamino]-N-hydroxy-2-(tetrahydro-2H-pyran-4-yl) acetamide, in complex with LF, prevented the zinc-dependent metalloprotease activity of LF and led to protection in various animal models55. The inhibitor bound within the active site of the LF subunit, effectively blocking activation. Crystallographic and pharmacological methods have been used to characterize 6 of 19 small-molecule inhibitors of LF56. These 19 compounds were identified on the basis of structural information and docking predictions from the US National Cancer Institute chemical repository. In contrast to other inhibitors that targeted the active binding pocket of the LF subunit, cisplatin did not inhibit the proteolytic activity of LF but prevented the cleavage of the mitogen-activated protein kinases (MAPKs) (Fig. 1). MAPKs are the major signalling molecules in human macrophages and neutrophils. MAPK activation results in the secretion of cytokines and the development of the adaptive immune response58. By inhibiting MAPKs in the macrophage, anthrax toxin prevents the exacerbation of the immune response that would occur following infection, by blocking both the innate and adaptive immune response and initiating an apoptotic pathway in the cell, resulting in cell death58.
Shiga toxin is a deadly toxin from EHEC that causes high morbidity and mortality during diarrhoeal outbreaks59,60,61. During EHEC infection, Shiga toxin is secreted into the lumen of the gastrointestinal tract, translocated across the intestinal epithelium and enters the systemic circulation. It is responsible for bloody diarrhoea and haemolytic uraemic syndrome (HUS), which leads to kidney and neurological sequelae59,60,61. The B subunit of the toxin binds specifically to the Gb3 glycolipid, which is present at a high density on renal tissue62 (Fig. 1). Gb3 molecular mimics that competitively inhibited the binding of Shiga toxin to its receptor Gb3, known as Synsorb-Pk, were developed in the late 1990s and early 2000s by a company named Synsorb, and were shown to be effective in removing free toxin from the gastrointestinal lumen, decreasing the possibility of sequelae in the human kidney and central nervous system63,64. The other advantage of this strategy is that the toxin bound to Synsorb-Pk is excreted from the body63. This prevents it from causing further damage, either inside or outside the body, as it is bound to its receptor. Although these initial studies demonstrated binding and removal of the toxin, there was not a significant improvement in the clinical outcome of HUS, showing that further studies are necessary. The commercial development of Synsorb-Pk has ceased but the drug remains one of the only treatments for EHEC.
Adhesion inhibitors. As discussed above, many bacteria, such as UPEC, use a binding structure known as a pilus to adhere to the host cell. Treatment with pilicides has been shown to decrease the efficiency of colonization of UPEC isolates in the urinary tract65. Pilicides are mimics of the normal pilin subunits that, when incorporated into the growing pilus, prevent elongation and the formation of a functional pilus. Small-molecule inhibitors of P pilus biogenesis, which belong to the class of bicyclic 2-pyridones, have also been identified66. A library of bicyclic 2-pyridones that inhibit the biogenesis of the type I secretion system and the P pilus of UPEC has been rationally designed67. In a mouse model of infection, the pilicides decreased the bacterial adhesion to the bladder and effectively prevented infection in the higher structures of the urogenital tract65,68. Inhibition of pilus biogenesis is a promising strategy to prevent adhesion of bacterial pathogens (Fig. 2).
Additional adhesion inhibitors could be similar to the synthetic receptor analogues used in the toxin treatments described above or direct inhibitors of adhesion. However, these are much more difficult to manipulate owing to the physical forces encountered at the mucosal membranes to which many pathogens bind.
Inhibitors of specialized bacterial secretory systems. Although there are many different types of TTSS, there are a limited number of ways to inhibit them. TTSS inhibitors prevent the assembly of the TTSS, inhibit the interaction with the eukaryotic host or inhibit the secretion of the effectors (Fig. 3).
Numerous small-molecule inhibitors of bacterial TTSS have been identified using high-throughput screening (HTS) of small-molecule libraries. Chlamydia spp. are obligate intracellular bacteria. The bacteria encode a TTSS that secretes peptides into the inclusion membrane of the infected host cell and is essential for intracellular survival of this pathogen69. A chemical class of small-molecule inhibitors known as acylated hydrazones of salicylaldehydes were shown to specifically block secretion through the TTSS of Chlamydia spp.70. In addition to preventing the TTSS-mediated secretion, these compounds also impaired the developmental stages of Chlamydia spp., locking the bacteria into one developmental phase and preventing the progression of disease. An additional study71 identified a separate class of molecules, including INP0400, which blocked the secretion systems of Chlamydia trachomatis and Yersinia pseudotuberculosis. Treatment with INP0400 blocked TTSS secretion at defined stages in the development of C. trachomatis and prevented the differentiation of Chlamydial developmental stages and virulence. Neither of these studies directly identified the precise mechanism and/or molecular target of TTSS inhibition by these small molecules.
Yersinia spp. virulence is linked to the secretion of effectors known as Yops (Yersinia outer-membrane proteins), which are secreted by the plasmid-encoded Ysc (Yersinia secretion system) TTSS. A ∼9,400 compound library was screened using a transcription reporter system involving yopE conjugated to luxAB, which encodes luciferase and provides the assay readout. Compounds 1–4 (Fig. 6; Table 2) were identified that were inhibitory and specific for TTSS72. Enteric pathogens infect the human gastrointestinal tract and many use a TTSS during infection for delivery of secreted effectors. Small-molecule screens have been performed for TTSS inhibitors of enteropathogenic E. coli73 (compounds containing salicylideaniline structures), Shigella spp.74 (compounds INP0400 and INP0406), Salmonella enterica serovar Typhimurium, Pseudomonas syringae and Yersinia enterocolytica (2-imino-5-arylidene thiazolidinone)75,76. Unique classes of TTSS inhibitors were identified for each species except Shigella, for which the inhibitors were of the same class of molecules that inhibited Chlamydia spp. — the salicylaldehydes (Figs 3,6; Table 2).
As evidenced in this section, substantial effort has focused on the attenuation of virulence by inhibiting the TTSS-mediated transfer of effectors into eukaryotic cells. However, although screens have identified a number of inhibitors, in general the precise mechanism of inhibition and molecular targets have yet to be identified.
Inhibitors of organism-specific virulence gene expression. Vibrio cholerae is responsible for some of the largest episodic outbreaks (pandemics) of diarrhoeal disease. Consequently, expression of virulence genes has been studied in detail in this organism77. Regulation of virulence in V. cholerae begins at the membrane with ToxRS and TcpPH, which in concert control expression of the transcription factor ToxT. ToxT dimerizes and directly activates expression of two crucial virulence factors: the cholera toxin and the toxin co-regulated pilus (TCP)77. Recently, a small-molecule screen of 50,000 compounds identified 15 compounds that inhibited virulence gene expression, but did not decrease growth nor kill the bacterium9,78. One compound, 4-[N-(1,8-naphthalimide)]-n-butyric acid (also known as virstatin), was shown to directly inhibit dimerization of ToxT, which is essential for ToxT-dependent transcriptional regulation and subsequent expression of cholera toxin and TCP. In vivo characterization in an infant-mouse model of infection showed reduced cholerae infectivity with 50 μM virstatin. This compound provides promise for the treatment of individuals in cholerae-endemic areas (Fig. 5A).
Inhibitors of organism-specific cell-to-cell signalling. Several strategies have been developed to inhibit the quorum-sensing systems that are responsible for mediating cell-to-cell signalling, downstream virulence gene expression and biofilm formation. Many strategies use small molecules identified through HTS of chemical libraries, or modelled antagonists based on signal and/or receptor molecular structure79,80,81. Other strategies use enzymes that degrade the quorum-sensing signal and have been referred to as quorum quenching82. Some bacteria, such as Bacillus spp., express lactonases that degrade the homoserine ring of AHLs82. Transgenic plants expressing the Bacillus lactonase show resistance to quorum sensing-dependent bacterial infection82.
Small-molecule quorum-sensing inhibitors can be either natural or synthetic compounds. The marine macroalga Delisea pulchra blocks virulence gene expression that is dependent on quorum sensing by producing a halogenated furanone that acts as a competitive inhibitor of the AHL-based quorum-sensing system of the bacterium83. C-30, a derivative of this furanone, was shown to inhibit quorum sensing and render Pseudomonas biofilms susceptible to clearance by detergent and antibiotics84,85. Non-biased HTS also produced a number of candidates for the inhibition of quorum sensing. HTS of a library of 200,000 small compounds identified two LasRI-based quorum-sensing inhibitors86: PD12 (a tetrazole with a 12-carbon alkyl tail), and V-06-018 (a phenyl ring with a 12-carbon alkyl tail). Further studies identified a library of inhibitors of C12-HSL–LasR signalling79,80,81. Although these molecules show promising in vitro results in preventing pathogenesis, their in vivo activity in animal models remains to be assessed.
Effective treatment of Gram-positive species, such as S. aureus, is now a major challenge owing to the rise of MRSA as well as other multidrug-resistant strains87,88. These bacterial species often use modified peptides to achieve cell-to-cell signalling89. The Agr quorum-sensing system is a key regulator of virulence gene expression in S. aureus90. It consists of an autoinducing peptide (AIP) that binds to and activates AgrC — a membrane-bound histidine sensor kinase and the main environmental sensory system in prokaryotic cells — which in turn regulates expression of virulence genes in S. aureus91. Four AIPs have been identified in S. aureus, which vary in amino-acid sequence. They have been shown to selectively bind to their cognate AgrC receptors and are naturally inhibited by quorum sensing when AIPs bind to non-cognate receptors92. It has been proposed that identifying or synthesizing competing analogues to AIPs will inhibit cell-to-cell signalling and could provide an alternative treatment to S. aureus infections92 (Fig. 4B).
These strategies to inhibit signalling are specific to the pathogen of interest and are not applicable to other, unrelated pathogens.
Broad-spectrum inhibitors of virulence and signalling. Although inhibition of virulence mediated by cell-to-cell signalling has been mainly approached in an organism-specific manner, a more recent study has targeted the common signalling pathway that begins with the membrane-bound QseC histidine sensor kinase5. QseC homologues are present in at least 25 important human and plant pathogens5, and qseC mutants of EHEC5,45, S. typhimurium5,46 and F. tularensis47 have been shown to be attenuated in animal models of infection. QseC increases its autophosphorylation in response to the bacterial quorum-sensing AI-3 and/or the host signals adrenaline and/or noradrenaline45, initiating a complex phosphorylation cascade in the bacterial cell that regulates the expression of virulence genes5,45,48. Histidine sensor kinases are ideal therapeutic targets as they are absent in mammalian cells.
HTS of a library of 150,000 small organic compounds led to the identification of a lead structure, LED209 (N-phenyl-4-[[(phenylamino)thioxomethyl]amino]-benzenesulphonamide), which blocks the binding of signalling molecules (AI-3, adrenaline and noradrenaline) to QseC. This prevents the autophosphorylation of QseC and consequently inhibits QseC-mediated activation of the expression of virulence genes5 (Fig. 4C). LED209 decreased virulence of EHEC in in vitro systems, whereas in vivo studies (in animals) did not show a statistically significant decrease in infection. It is thought this failure of LED209 in the EHEC animal model is due to the rapid systemic absorption of the compound into the peripheral circulatory system.
Based on homology, QseC was shown to be present in many bacterial species including Salmonella and Francisella species. In vitro characterization of LED209 in these other species showed that the compound caused a decrease in the expression of virulence genes and a congruent decrease in virulence in models of infection5. Although each of these pathogens infects a different location in the host and has variable cell tropism, bacterial infectivity was decreased with nanomolar or picomolar doses of LED209. These are at least 10 orders of magnitude lower than all previously characterized virulence inhibitory compounds. This study revealed that QseC is an attractive target for novel therapeutics. As it is present in many important animal and plant pathogens, drugs that target this sensor kinase would potentially have a broad spectrum of activity. In addition, the QseC-dependent inter-kingdom signalling system does not directly influence bacterial growth; therefore, inhibition of this signalling pathway may not exert strong selective pressure towards development of drug resistance. LED290 provides proof of principle that a strategy to develop drugs that interfere with inter-kingdom chemical signalling is a feasible approach to developing novel therapeutics to combat bacterial infections.
Potential and limitations
As discussed above, the greatest advantage to targeting the virulence factors of pathogens is that there is milder evolutionary pressure for the development of resistance. Therefore, an optimally designed antimicrobial agent should not inhibit growth and only diminish the effects of the virulence traits. This will prevent the bacteria from colonizing the host, instead causing them to pass through the host without detrimental effects. Although the normal host immune response might be able to resolve any residual infection after treatment with an inhibitor that specifically targets virulence, such therapies could be coupled with more traditional antibiotics to increase their efficacy and potentially decrease the development of resistance to these existing antimicrobials. An optimal therapeutic agent would target virulence factors or signalling pathways that are present in several pathogens, as is the case for the QseC signalling pathway5, allowing for the development of a broader-spectrum drug. The use of broad-spectrum therapies provides the possibility to use one compound to treat many infections. Narrow-spectrum therapies would be most effective if rapid diagnostics were available to identify the pathogen responsible for the disease.
Safety is always a concern when introducing a potential new therapeutic. Given that these therapies are targeted to virulence pathways or factors that exist exclusively in pathogens, it is likely that there will be less interference with mammalian signalling pathways — and therefore reduced toxicity and less severe side effects — than with therapies that affect targets which are also expressed by the host. However, although inhibitors that reduce virulence are targeted to virulence traits in pathogenic isolates, they might also target similar pathways or factors in the normal microbiota. In the case of QseC, the receptor is present not only in pathogens but also in commensal species of the gastrointestinal microbiota5,93. Although no adverse effects were observed in mice treated with LED209, this does not rule out potential detrimental effects on the gastrointestinal microbiota5. It is unclear what effects these types of therapeutics will have on the species composition of these commensal communities. The removal of species, or alteration of the balance of species in a community as complex as the gastrointestinal microbiota, could result in detrimental effects for the host.
Although it is assumed that anti-virulence drugs will engender a milder evolutionary pressure for the development of antimicrobial resistance, it is unlikely that resistance will not develop over time. The rapid rate of bacterial evolution enables a pathogen to change to an alternative route to the one being targeted to achieve infection. Vigilance and basic research in the understanding of the key processes associated with bacterial virulence are crucial to the development of novel and effective therapeutics.
As none of these anti-virulence strategies has been applied in a broad-scale clinical trial, it is difficult to determine the potential side effects. However, if the drugs are rationally designed, they should target only the pathogenic bacteria and spare the harmful side effects that are commonly observed with broad-spectrum antibiotics. Nevertheless, it is possible that these drugs may produce secondary metabolites that may impair the growth of the bacteria or have detrimental effects in the host. The best approach remains to follow the rules of rational drug design and rigorously test these compounds before they are released onto the market.
Concluding remarks
The avid research on the development of anti-virulence therapeutics indicates a paradigm shift in the understanding of bacterial pathogens and their interactions with the host and environment. It is now appreciated that bacteria have important beneficial interactions with the host, and that they live in equilibrium in polymicrobial communities. If virulence can be controlled, it is hoped that the host immune system will be capable of overcoming any infections that the therapeutic is not. In cases in which this does not occur, anti-virulence therapies could be used synergistically with traditional antimicrobials, allowing less of each agent to be used. Additionally, with less of the therapeutic agent there will be less of an opportunity for the pathogen to develop resistance, increasing the time for which both types of therapy can be used.
However, several questions remain to be answered. Is it possible to eradicate the pathogen without affecting the microbiota? Should a mono-species infection be treated differently from a polymicrobial infection? Because anti-virulence drugs will not kill the bacteria, will they be more effective than conventional antimicrobials and will they delay the development of drug-resistance? The further development of novel anti-virulence therapies will require continued investigations in basic science to further understand the bacterial virulence strategies, signalling pathways and potential ways to exploit them.
References
Staskawicz, B. J., Mudgett, M. B., Dangl, J. L. & Galan, J. E. Common and contrasting themes of plant and animal diseases. Science 292, 2285–2289 (2001).
Walsh, C. Antibiotics: Actions, Origins, Resistance (ASM Press, Washington, D. C., 2003).
Werner, G., Strommenger, B. & Witte, W. Acquired vancomycin resistance in clinically relevant pathogens. Future Microbiol. 3, 547–562 (2008).
Hanson, M. R. & Chung, C. L. Antibiotic selection for MRSA: case presentations and review of the literature. J. Drugs Dermatol. 8, 281–286 (2009).
Rasko, D. A. et al. Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080 (2008). The first study to show that inhibitors of bacteria and bacterial–host chemical signalling worked in animal models and blocked pathogenesis of three important human pathogens.
Rumbaugh, K. P. Convergence of hormones and autoinducers at the host/pathogen interface. Anal. Bioanal. Chem. 387, 425–435 (2007).
Paul, M. & Leibovici, L. Combination antimicrobial treatment versus monotherapy: the contribution of meta-analyses. Infect. Dis. Clin. North Am. 23, 277–293 (2009).
Cegelski, L., Marshall, G. R., Eldridge, G. R. & Hultgren, S. J. The biology and future prospects of antivirulence therapies. Nature Rev. Microbiol. 6, 17–27 (2008).
Hung, D. T., Shakhnovich, E. A., Pierson, E. & Mekalanos, J. J. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310, 670–674 (2005). The first article showing that a targeted anti-virulence approach could prevent cholerae disease in animal models.
Shrivastava, R. & Miller, J. F. Virulence factor secretion and translocation by Bordetella species. Curr. Opin. Microbiol. 12, 88–93 (2009).
Hedge, D. D., Strain, J. D., Heins, J. R. & Farver, D. K. New advances in the treatment of Clostridium difficile infection (CDI). Ther. Clin. Risk Manag. 4, 949–964 (2008).
O'Brien, A. D. et al. Shigellosis and Escherichia coli diarrhea: relative importance of invasive and toxigenic mechanisms. Am. J. Clin. Nutr. 32, 229–233 (1979).
Schiavo, G. & van der Goot, F. G. The bacterial toxin toolkit. Nature Rev. Mol. Cell Biol. 2, 530–537 (2001).
Kaper, J. B., Nataro, J. P. & Mobley, H. L. Pathogenic. Escherichia coli. Nature Rev. Microbiol. 2, 123–140 (2004).
Abrami, L., Liu, S., Cosson, P., Leppla, S. H. & van der Goot, F. G. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J. Cell Biol. 160, 321–328 (2003).
Keim, P., Mock, M., Young, J. & Koehler, T. M. The International Bacillus anthracis, B. cereus, and B. thuringiensis Conference, “Bacillus-ACT05”. J. Bacteriol. 188, 3433–3441 (2006).
Ofek, I., Hasty, D. L. & Doyle, R. J. Bacterial Adhesion to Animal Cells and Tissues (ASM Press, Washington DC, 2003).
Proft, T. & Baker, E. N. Pili in Gram-negative and Gram-positive bacteria — structure, assembly and their role in disease. Cell. Mol. Life Sci. 66, 613–635 (2009).
Wright, K. J. & Hultgren, S. J. Sticky fibers and uropathogenesis: bacterial adhesins in the urinary tract. Future Microbiol. 1, 75–87 (2006).
Lane, M. C. & Mobley, H. L. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 72, 19–25 (2007).
Welch, R. A. et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic. Escherichia coli. Proc. Natl Acad. Sci. USA 99, 17020–17024 (2002).
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999).
Singh, P. K. et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764 (2000). The first study to show that bacterial quorum-sensing signals are present in the lungs of patients with cystic fibrosis.
Olson, M. E., Ceri, H., Morck, D. W., Buret, A. G. & Read, R. R. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 66, 86–92 (2002).
Galan, J. E. & Cossart, P. Host-pathogen interactions: a diversity of themes, a variety of molecular machines. Curr. Opin. Microbiol. 8, 1–3 (2005).
Filloux, A., Hachani, A. & Bleves, S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 (2008).
Keyser, P., Elofsson, M., Rosell, S. & Wolf-Watz, H. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264, 17–29 (2008).
Stavrinides, J., McCann, H. C. & Guttman, D. S. Host–pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 10, 285–292 (2008).
Filloux, A. The type VI secretion system: a tubular story. EMBO J. 28, 309–310 (2009).
DiRita, V. J. et al. Virulence gene regulation inside and outside. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 657–665 (2000).
Lowery, C. A., Dickerson, T. J. & Janda, K. D. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 37, 1337–1346 (2008).
Hughes, D. T. & Sperandio, V. Inter-kingdom signalling: communication between bacteria and their hosts. Nature Rev. Microbiol. 6, 111–120 (2008).
Fuqua, W. C., Winans, S. C. & Greenberg, E. P. Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994). This article coined the term quorum sensing to describe bacterial chemical communication.
Engebrecht, J., Nealson, K. & Silverman, M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781 (1983).
Nealson, K. H., Platt, T. & Hastings, J. W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322 (1970).
Gooderham, W. J. & Hancock, R. E. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33, 279–294 (2009).
Pearson, J. P. et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl Acad. Sci. USA 91, 197–201 (1994).
Pearson, J. P., Pesci, E. C. & Iglewski, B. H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179, 5756–5767 (1997).
Brint, J. M. & Ohman, D. E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR–RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR–LuxI family. J. Bacteriol. 177, 7155–7163 (1995).
Pesci, E. C. et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 96, 11229–11234 (1999).
Wilson, R. et al. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56, 2515–2517 (1988).
Wade, D. S. et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187, 4372–4380 (2005).
Edrington, T. S. et al. Acyl-homoserine-lactone autoinducer in the gastrointesinal tract of feedlot cattle and correlation to season, E. Coli O157:H7 prevalence, and diet. Curr. Microbiol. 58, 227–232 (2008).
Mitrophanov, A. Y. & Groisman, E. A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611 (2008).
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C. & Sperandio, V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl Acad. Sci. USA 103, 10420–10425 (2006). This paper describes the identification of the first bacterial adrenergic receptor.
Bearson, B. L. & Bearson, S. M. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278 (2007).
Weiss, D. S. et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl Acad. Sci. USA 104, 6037–6042 (2007).
Clarke, M. B. & Sperandio, V. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57, 1734–1749 (2005).
Welkos, S. L. & Friedlander, A. M. Comparative safety and efficacy against. Bacillus anthracis of protective antigen and live vaccines in mice. Microb. Pathog. 5, 127–139 (1988).
Flynn, E. R., Bradley, K. N., Muir, T. C. & McCarron, J. G. Functionally separate intracellular Ca2+ stores in smooth muscle. J. Biol. Chem. 276, 36411–36418 (2001).
Bradley, K. A. & Young, J. A. Anthrax toxin receptor proteins. Biochem. Pharmacol. 65, 309–314 (2003).
Scobie, H. M. et al. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J. Infect. Dis. 192, 1047–1051 (2005).
Chen, Z. et al. Efficient neutralization of anthrax toxin by chimpanzee monoclonal antibodies against protective antigen. J. Infect. Dis. 193, 625–633 (2006).
Karginov, V. A., Nestorovich, E. M., Moayeri, M., Leppla, S. H. & Bezrukov, S. M. Blocking anthrax lethal toxin at the protective antigen channel by using structure-inspired drug design. Proc. Natl Acad. Sci. USA 102, 15075–15080 (2005).
Shoop, W. L. et al. Anthrax lethal factor inhibition. Proc. Natl Acad. Sci. USA 102, 7958–7963 (2005).
Panchal, R. G. et al. Identification of small molecule inhibitors of anthrax lethal factor. Nature Struct. Mol. Biol. 11, 67–72 (2004).
Moayeri, M., Wiggins, J. F., Lindeman, R. E. & Leppla, S. H. Cisplatin inhibition of anthrax lethal toxin. Antimicrob. Agents Chemother. 50, 2658–2665 (2006).
Turk, B. E. Manipulation of host signalling pathways by anthrax toxins. Biochem. J. 402, 405–417 (2007).
Greener, M. How Escherichia coli kills. Mol. Med. Today 6, 411 (2000).
Karmali, M. A., Petric, M., Lim, C., Fleming, P. C. & Steele, B. T. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet 2, 1299–1300 (1983).
Karmali, M. A., Steele, B. T., Petric, M. & Lim, C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1, 619–620 (1983).
Boyd, B. & Lingwood, C. Verotoxin receptor glycolipid in human renal tissue. Nephron 51, 207–210 (1989).
Armstrong, G. D. et al. A phase I study of chemically synthesized verotoxin (Shiga-like toxin) Pk-trisaccharide receptors attached to chromosorb for preventing hemolytic-uremic syndrome. J. Infect. Dis. 171, 1042–1045 (1995). This paper describes the development of Shiga toxin receptor mimics as anti-virulence therapies.
Trachtman, H. et al. Effect of an oral Shiga toxin-binding agent on diarrhea-associated hemolytic uremic syndrome in children: a randomized controlled trial. JAMA 290, 1337–1344 (2003).
Svensson, A. et al. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. Chembiochem 2, 915–918 (2001).
Berg, V. et al. Design, synthesis and evaluation of peptidomimetics based on substituted bicyclic 2-pyridones-targeting virulence of uropathogenic E. coli. Bioorg. Med. Chem. 14, 7563–7581 (2006).
Pinkner, J. S. et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl Acad. Sci. USA 103, 17897–17902 (2006). This study describes the development of inhibitors of adhesins.
Svensson, M. et al. Carbohydrate receptor depletion as an antimicrobial strategy for prevention of urinary tract infection. J. Infect. Dis. 183, S70–S73 (2001).
Hueck, C. J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 (1998). This study describes the development of inhibitors of bacterial TTSS.
Bailey, L. et al. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581, 587–595 (2007).
Muschiol, S. et al. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl Acad. Sci. USA 103, 14566–14571 (2006).
Kauppi, A. M., Nordfelth, R., Uvell, H., Wolf-Watz, H. & Elofsson, M. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10, 241–249 (2003).
Gauthier, A. et al. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49, 4101–4109 (2005).
Veenendaal, A. K., Sundin, C. & Blocker, A. J. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191, 563–570 (2009).
Felise, H. B. et al. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4, 325–336 (2008).
Kline, T. et al. Substituted 2-imino-5-arylidenethiazolidin-4-one inhibitors of bacterial type III secretion. J. Med. Chem. 51, 7065–7074 (2008).
Matson, J. S., Withey, J. H. & DiRita, V. J. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75, 5542–5549 (2007).
Shakhnovich, E. A., Hung, D. T., Pierson, E., Lee, K. & Mekalanos, J. J. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc. Natl Acad. Sci. USA 104, 2372–2377 (2007).
Smith, K. M., Bu, Y. & Suga, H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 10, 563–571 (2003). One of the first HTS screens to identify compounds that inhibit bacterial quorum sensing.
Smith, K. M., Bu, Y. & Suga, H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 10, 81–89 (2003).
Suga, H. & Smith, K. M. Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr. Opin. Chem. Biol. 7, 586–591 (2003).
Dong, Y. H. et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817 (2001).
Rasmussen, T. B. et al. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146, 3237–3244 (2000).
Hentzer, M. et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815 (2003).
Manefield, M. et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148, 1119–1127 (2002).
Muh, U. et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50, 3674–3679 (2006).
Weinstein, R. A. Controlling antimicrobial resistance in hospitals: infection control and use of antibiotics. Emerg. Infect. Dis. 7, 188–192 (2001).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Novick, R. P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 (2003).
Novick, R. P. & Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008).
George, E. A. & Muir, T. W. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem 8, 847–855 (2007).
Wright, J. S. 3rd, Jin, R. & Novick, R. P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl Acad. Sci. USA 102, 1691–1696 (2005).
Walters, M., Sircili, M. P. & Sperandio, V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188, 5668–5681 (2006).
Acknowledgements
Work in the Sperandio laboratory is supported by the National Institutes of Health and Burroughs Wellcome Fund. The authors apologize to the numerous investigators whose manuscripts could not be cited owing to space constrains.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
V.S. is a member of the scientific advisory board of OMM Scientific.
Glossary
- Anti-virulence drug
-
A drug that targets bacterial virulence but is not bacteriostatic (growth inhibiting) or bacteriocidal (bacteria killing).
- Adhesins
-
A broad group of structures produced by bacteria that facilitate bacterial colonization of the host by promoting binding to host cells.
- Toxins
-
Bacterial proteins that have detrimental effects on host cells.
- Effector
-
A bacterial protein that functions as a molecular mimic of a eukaryotic protein.
- Quorum sensing
-
Cell-to-cell chemical signalling among bacterial cells, analogous to hormonal signalling in mammalian systems.
- Histidine sensor kinase
-
A bacterial sensor of environmental cues.
Rights and permissions
About this article
Cite this article
Rasko, D., Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9, 117–128 (2010). https://doi.org/10.1038/nrd3013
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd3013
This article is cited by
-
Ziziphus spina-christi (L.) Willd. Leaves Extract Affecting agr Quorum Sensing System in Staphylococcus aureus
Arabian Journal for Science and Engineering (2024)
-
Antibiofilm and antivirulence activities of laminarin-gold nanoparticles in standard and host-mimicking media
Applied Microbiology and Biotechnology (2024)
-
Interception of Epoxide ring to quorum sensing system in Enterococcus faecalis and Staphylococcus aureus
AMB Express (2023)
-
It is the time for quorum sensing inhibition as alternative strategy of antimicrobial therapy
Cell Communication and Signaling (2023)
-
Development of a smart pH-responsive nano-polymer drug, 2-methoxy-4-vinylphenol conjugate against the intestinal pathogen, Vibrio cholerae
Scientific Reports (2023)