Key Points
-
WD40 repeat (WDR) domains are β-propeller domains that act as protein interaction scaffolds in multiprotein complexes.
-
WDR domain-containing proteins comprise one of the largest protein families in humans and are involved in a diverse array of cellular networks, many of which are disease-associated.
-
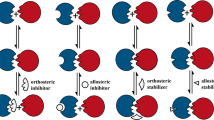
The central cavity of the WDR domain is structurally dynamic and diverse. Potent, selective inhibitors target the central cavity of WD repeat-containing protein 5 (WDR5) and EED, two proteins involved in chromatin complexes. An EED inhibitor has entered clinical trials in oncology.
-
Targeting WDR domains that bind to other proteins that have so far proved undruggable, such as components of the ubiquitin–proteasome system, is a promising but underexplored strategy.
-
The cellular implications of pharmacologically targeting WDR domains that are involved in multiple protein complexes are still poorly understood.
Abstract
Antagonism of protein–protein interactions (PPIs) with small molecules is becoming more feasible as a therapeutic approach. Successful PPI inhibitors tend to target proteins containing deep peptide-binding grooves or pockets rather than the more common large, flat protein interaction surfaces. Here, we review one of the most abundant PPI domains in the human proteome, the WD40 repeat (WDR) domain, which has a central peptide-binding pocket and is a member of the β-propeller domain-containing protein family. Recently, two WDR domain-containing proteins, WDR5 and EED, as well as other β-propeller domains have been successfully targeted by potent, specific, cell-active, drug-like chemical probes. Could WDR domains be a novel target class for drug discovery? Although the research is at an early stage and therefore not clinically validated, cautious optimism is justified, as WDR domain-containing proteins are involved in multiple disease-associated pathways. The druggability and structural diversity of WDR domain binding pockets suggest that understanding how to target this prevalent domain class will open up areas of disease biology that have so far resisted drug discovery efforts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deeks, E. D. Venetoclax: first global approval. Drugs 76, 979–987 (2016).
Roberts, A. W. et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 374, 311–322 (2016).
Shangary, S. & Wang, S. Small-molecule inhibitors of the MDM2–p53 protein–protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 49, 223–241 (2009).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Shi, J. & Vakoc, C. R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Scott, D. E., Bayly, A. R., Abell, C. & Skidmore, J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 (2016).
Niesen, F. H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007).
Annis, D. A., Nickbarg, E., Yang, X., Ziebell, M. R. & Whitehurst, C. E. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr. Opin. Chem. Biol. 11, 518–526 (2007).
Clark, M. A. et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654 (2009).
Pearce, N. M. et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 8, 15123 (2017).
Lea, W. A. & Simeonov, A. Fluorescence polarization assays in small molecule screening. Expert Opin. Drug Discov. 6, 17–32 (2011).
Juhasz, T., Szeltner, Z., Fulop, V. & Polgar, L. Unclosed β-propellers display stable structures: implications for substrate access to the active site of prolyl oligopeptidase. J. Mol. Biol. 346, 907–917 (2005).
Fulop, V., Bocskei, Z. & Polgar, L. Prolyl oligopeptidase: an unusual β-propeller domain regulates proteolysis. Cell 94, 161–170 (1998).
Stirnimann, C. U., Petsalaki, E., Russell, R. B. & Muller, C. W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574 (2010). This article discusses the prevalence of WDR domains in protein networks and the structural arrangement of WDR domain-containing complexes.
Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Migliori, V. et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 19, 136–144 (2012).
Hao, B., Oehlmann, S., Sowa, M. E., Harper, J. W. & Pavletich, N. P. Structure of a Fbw7–Skp1–cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007).
Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 (2003).
Wang, Y. et al. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res. 43, D339–D344 (2015).
Wang, Y., Jiang, F., Zhuo, Z., Wu, X. H. & Wu, Y. D. A method for WD40 repeat detection and secondary structure prediction. PLoS ONE 8, e65705 (2013).
Chen, C. K., Chan, N. L. & Wang, A. H. The many blades of the β-propeller proteins: conserved but versatile. Trends Biochem. Sci. 36, 553–561 (2011).
Croft, D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 (2014).
Grebien, F. et al. Pharmacological targeting of the Wdr5–MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 11, 571–578 (2015). This article reports on a nanomolar inhibitor of a WDR domain.
He, Y. et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 13, 389–395 (2017).
Qi, W. et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 13, 381–388 (2017). Refs 26 and 27 describe inhibitors that target a WDR domain of PRC2 and are active against cancer cells that are resistant to catalytic inhibitors.
Davis, R. J., Welcker, M. & Clurman, B. E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26, 455–464 (2014).
Laskowski, R. A. et al. Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Hum. Mol. Genet. 25, 927–935 (2016).
Dawson, M. A. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 (2017).
Pfister, S. X. & Ashworth, A. Marked for death: targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 16, 241–263 (2017).
Shortt, J., Ott, C. J., Johnstone, R. W. & Bradner, J. E. A chemical probe toolbox for dissecting the cancer epigenome. Nat. Rev. Cancer 17, 160–183 (2017).
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Zhu, J. et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 (2015).
Thiel, A. T. et al. MLL–AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 17, 148–159 (2010).
Cao, F. et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 53, 247–261 (2014).
Li, Y. et al. Structural basis for activity regulation of MLL family methyltransferases. Nature 530, 447–452 (2016).
Garapaty-Rao, S. et al. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem. Biol. 20, 1329–1339 (2013).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Konze, K. D. et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8, 1324–1334 (2013).
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 8, 890–896 (2012).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Kim, W. et al. Targeted disruption of the EZH2–EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 9, 643–650 (2013).
Wolffe, A. P., Urnov, F. D. & Guschin, D. Co-repressor complexes and remodelling chromatin for repression. Biochem. Soc. Trans. 28, 379–386 (2000).
Kitange, G. J. et al. Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 14, 2587–2598 (2016).
Chan-Penebre, E. et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437 (2015).
Kryukov, G. V. et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 (2016).
Mavrakis, K. J. et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213 (2016).
Chen, H., Lorton, B., Gupta, V. & Shechter, D. A. TGFβ–PRMT5–MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36, 373–386 (2017).
Peters, J. M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656 (2006).
Kidokoro, T. et al. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 27, 1562–1571 (2008).
Mao, D. D. et al. A CDC20–APC/SOX2 signaling axis regulates human glioblastoma stem-like cells. Cell Rep. 11, 1809–1821 (2015).
Xie, Q. et al. CDC20 maintains tumor initiating cells. Oncotarget 6, 13241–13254 (2015).
Marucci, G. et al. Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Arch. 453, 599–609 (2008).
Van Roosbroeck, K. et al. JAK2 rearrangements, including the novel SEC31A–JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood 117, 4056–4064 (2011).
Bedwell, C. et al. Cytogenetically complex SEC31A–ALK fusions are recurrent in ALK-positive large B-cell lymphomas. Haematologica 96, 343–346 (2011).
Van Roosbroeck, K. et al. ALK-positive large B-cell lymphomas with cryptic SEC31A–ALK and NPM1–ALK fusions. Haematologica 95, 509–513 (2010).
Xia, B. et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39, 159–161 (2007).
Reid, S. et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 39, 162–164 (2007).
Rahman, N. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39, 165–167 (2007).
Xia, B. et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 (2006).
Erkko, H. et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446, 316–319 (2007).
Smith, M. A. et al. Initial testing (stage 1) of the PARP inhibitor BMN 673 by the pediatric preclinical testing program: PALB2 mutation predicts exceptional in vivo response to BMN 673. Pediatr. Blood Cancer 62, 91–98 (2015).
Martini, E., Roche, D. M., Marheineke, K., Verreault, A. & Almouzni, G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143, 563–575 (1998).
Cheloufi, S. et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015).
Zhang, H. et al. MLL1 inhibition reprograms epiblast stem cells to naive pluripotency. Cell Stem Cell 18, 481–494 (2016).
Tanaka, T. et al. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165, 709–721 (2004).
Haverfield, E. V., Whited, A. J., Petras, K. S., Dobyns, W. B. & Das, S. Intragenic deletions and duplications of the LIS1 and DCX genes: a major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur. J. Hum. Genet. 17, 911–918 (2009).
Reiner, O. et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature 364, 717–721 (1993).
Bi, W. et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 41, 168–177 (2009).
Healy, D. G. et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 7, 583–590 (2008).
Smith, W. W. et al. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 9, 1231–1233 (2006).
West, A. B. et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl Acad. Sci. USA 102, 16842–16847 (2005).
Li, T. et al. Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson's disease cell and mouse models. Hum. Mol. Genet. 23, 6212–6222 (2014).
Deng, X. et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 7, 203–205 (2011).
Lee, B. D. et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat. Med. 16, 998–1000 (2010).
Kett, L. R. et al. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 21, 890–899 (2012).
Jorgensen, N. D. et al.The WD40 domain is required for LRRK2 neurotoxicity. PLoS ONE 4, e8463 (2009).
Klenke, S., Kussmann, M. & Siffert, W. The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet. Genomics 21, 594–606 (2011).
Siffert, W. et al. Association of a human G-protein β3 subunit variant with hypertension. Nat. Genet. 18, 45–48 (1998).
Goldlust, I. S. et al. Mouse model implicates GNB3 duplication in a childhood obesity syndrome. Proc. Natl Acad. Sci. USA 110, 14990–14994 (2013).
Nilsson, J., Sengupta, J., Frank, J. & Nissen, P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5, 1137–1141 (2004).
McCahill, A., Warwicker, J., Bolger, G. B., Houslay, M. D. & Yarwood, S. J. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62, 1261–1273 (2002).
Boyle, W. J., Simonet, W. S. & Lacey, D. L. Osteoclast differentiation and activation. Nature 423, 337–342 (2003).
Lin, J., Lee, D., Choi, Y. & Lee, S. Y. The scaffold protein RACK1 mediates the RANKL-dependent activation of p38 MAPK in osteoclast precursors. Sci. Signal 8, ra54 (2015).
Majzoub, K. et al. RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095 (2014).
Kim, K. et al. VprBP has intrinsic kinase activity targeting histone H2A and represses gene transcription. Mol. Cell 52, 459–467 (2013).
Angers, S. et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443, 590–593 (2006).
Wu, Y. et al. The DDB1–DCAF1–Vpr–UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 23, 933–940 (2016).
Schwefel, D. et al. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505, 234–238 (2014).
Li, T., Chen, X., Garbutt, K. C., Zhou, P. & Zheng, N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124, 105–117 (2006).
Orlicky, S. et al. An allosteric inhibitor of substrate recognition by the SCFCdc4 ubiquitin ligase. Nat. Biotechnol. 28, 733–737 (2010). The inhibitor in this article binds at a side pocket of a WDR domain and allosterically occludes the central cavity.
Sackton, K. L. et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649 (2014).
Bolshan, Y. et al. Synthesis, optimization, and evaluation of novel small molecules as antagonists of WDR5–MLL interaction. ACS Med. Chem. Lett. 4, 353–357 (2013).
Getlik, M. et al. Structure-based optimization of a small molecule antagonist of the interaction between WD repeat-containing protein 5 (WDR5) and mixed-lineage leukemia 1 (MLL1). J. Med. Chem. 59, 2478–2496 (2016).
Senisterra, G. et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 449, 151–159 (2013).
Curtin, M. L. et al. SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding. Bioorg. Med. Chem. Lett. 27, 1576–1583 (2017).
Huang, Y. et al. Discovery of first-in-class, potent, and orally bioavailable embryonic ectoderm development (EED) inhibitor with robust anticancer efficacy. J. Med. Chem. 60, 2215–2226 (2017).
Li, L. et al. Discovery and molecular basis of a diverse set of polycomb repressive complex 2 inhibitors recognition by EED. PLoS ONE 12, e0169855 (2017).
Csizmok, V. et al. An allosteric conduit facilitates dynamic multisite substrate recognition by the SCFCdc4 ubiquitin ligase. Nat. Commun. 8, 13943 (2017).
Davies, T. G. et al. Monoacidic inhibitors of the kelch-like ech-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein–protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem. 59, 3991–4006 (2016). This article describes a fragment-based approach to target the central cavity of a β-propeller domain.
Kerry, P. S. et al. Structural basis for a class of nanomolar influenza A neuraminidase inhibitors. Sci. Rep. 3, 2871 (2013).
von Kleist, L. et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146, 471–484 (2011).
Korczynska, M., Mukhtar, T. A., Wright, G. D. & Berghuis, A. M. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc. Natl Acad. Sci. USA 104, 10388–10393 (2007).
Springer, T. A., Zhu, J. & Xiao, T. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3 . J. Cell Biol. 182, 791–800 (2008).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007). This study reports on a small molecule that activates a β-propeller domain.
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405 (2010).
Avdic, V. et al. Structural and biochemical insights into MLL1 core complex assembly. Structure 19, 101–108 (2011).
Baker, T. et al. Acquisition of a single EZH2 D1 domain mutation confers acquired resistance to EZH2-targeted inhibitors. Oncotarget 6, 32646–32655 (2015).
Gibaja, V. et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene 35, 558–566 (2016).
Edwards, A. M. et al. Preclinical target validation using patient-derived cells. Nat. Rev. Drug Discov. 14, 149–150 (2015).
Bunnage, M. E., Chekler, E. L. & Jones, L. H. Target validation using chemical probes. Nat. Chem. Biol. 9, 195–199 (2013).
Frye, S. V. The art of the chemical probe. Nat. Chem. Biol. 6, 159–161 (2010).
Kronke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001).
Saenz, D. T. et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia http://dx.doi.org/10.1038/leu.2016.393 (2017).
Uehara, T. et al. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat. Chem. Biol. (2017).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017). Refs 119 and 120 describe a WDR-containing protein as the target of a Protac small molecule, resulting in ubiquitylation and degradation of an oncoprotein.
Acknowledgements
The Structural Genomics Consortium (SGC) is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute (OGI-055), Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant no. 115766), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, São Paulo Research Foundation-FAPESP, Takeda and the Wellcome Trust. C.H.A. is supported by the Canadian Institute for Health Research (CIHR) (FRN-125792) and holds a Canada Research Chair in Structural Genomics. M.Ty. is supported by grants from the CIHR (MOP 126129), the Canadian Cancer Society Research Institute (703906), Genome Canada, Genome Quebec and National Institutes of Health (R01OD010929), and holds a Canada Research Chair in Systems and Synthetic Biology. The authors acknowledge Z.D. Wang for his contribution in generating the list of 361 WDR-containing proteins and C. Jakob for her careful review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.To. is an employee of AbbVie and owns AbbVie stock. M.S., M.Ty. and C.H.A. declare no competing interests.
Supplementary information
Supplementary information
Supplementary Tables (PDF 2958 kb)
Glossary
- Thermal stabilization
-
A thermodynamic property used to screen chemical libraries, and which relies on the fact that the melting point of a protein often increases upon the binding of a small molecule.
- Affinity selection–mass spectrometry
-
A screening technique whereby a protein is exposed to small molecules, receptor–ligand complexes are isolated and bound ligands are identified by mass spectrometry.
- DNA-encoded libraries
-
Chemical libraries in which each small molecule is linked to a unique DNA fragment that serves as a barcode. These typically very large libraries can be interrogated by affinity selection.
- High-throughput fragment screening
-
The screening of chemical libraries composed of chemical fragments (molecules smaller than 300 Da) typically at high concentration using direct binding rather than enzymatic assays.
- Fluorescence polarization
-
Fluorescence-based binding assay that relies on the fact that free molecules rotate faster than large molecular complexes.
- Apo state
-
The unbound state of a protein or other macromolecule, as opposed to the 'holo' state, when complexed to a ligand.
- Bidentate ligands
-
Compounds with two chemical groups carrying distinct functions, for example each binding to a different protein interaction partner.
Rights and permissions
About this article
Cite this article
Schapira, M., Tyers, M., Torrent, M. et al. WD40 repeat domain proteins: a novel target class?. Nat Rev Drug Discov 16, 773–786 (2017). https://doi.org/10.1038/nrd.2017.179
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2017.179
This article is cited by
-
WD repeat domain 76 predicts poor prognosis in lower grade glioma and provides an original target for immunotherapy
European Journal of Medical Research (2024)
-
Pocket Crafter: a 3D generative modeling based workflow for the rapid generation of hit molecules in drug discovery
Journal of Cheminformatics (2024)
-
Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites
Applied Microbiology and Biotechnology (2024)
-
Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF
Nature (2024)
-
DCAF1-based PROTACs with activity against clinically validated targets overcoming intrinsic- and acquired-degrader resistance
Nature Communications (2024)