Key Points
-
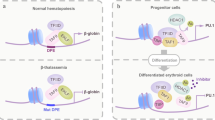
The heterodimeric transcription factor core-binding factors (CBFs) are comprised of the RUNX1 (also known as AML1, CBFA2 and PEBP2αB) and CBFβ subunits.
-
The RUNX1 subunit directly contacts DNA, and binding affinity for DNA is significantly increased by association with CBFβ, which does not contact DNA.
-
CBFs are involved in haematopoietic development. Knock-out mice indicate that both subunits of CBF are absolutely required for definitive haematopoiesis.
-
Correct CBF gene dosage is important during haematopoietic development. Subtle changes in the dosage of Runx1 can significantly affect the timing of stem-cell emergence and the number of committed progenitors, and cause alterations in T-cell development. Haploinsufficiency can contribute to inherited leukaemia syndromes, and some leukaemias seem to have amplification of the RUNX1 locus.
-
Chromosomal translocations target CBFs in human leukaemias. These include the RUNX1–ETO, CBFB–SMMHC and TEL–RUNX1 fusion genes. Biochemical and developmental studies indicate that these all encode dominant inhibitors of the native CBF complex (RUNX1–ETO, CBFβ–SMMHC and TEL–RUNX1), through recruitment of the nuclear corepressor complex.
-
These fusion genes contribute to the pathogenesis of leukaemia. Their expression seems to impair normal haematopoietic development, but is not sufficient to cause leukaemia. A two-hit model of disease pathogenesis for CBF leukaemias is presented.
-
CBF fusions, in general, confer a favourable prognosis in patients with leukaemia, although there is debate about the prognostic significance of the TEL–RUNX1 fusion in paediatric acute lymphoblastic leukaemia.
-
Therapeutic strategies for targeting CBF in leukaemia could include the use of histone-deacetylase inhibitors to disrupt the nuclear corepressor function that is mediated by the CBF fusions.
Abstract
Core-binding factors (CBFs) are a class of haematopoietic transcription factors that are crucial for the regulation of haematopoietic ontogeny, and are frequent targets of mutation and gene rearrangement in human leukaemia. So, what are the functions of CBFs during development, and what are the functional consequences of CBF mutations in leukaemia? Synergy between these convergent lines of enquiry has furthered our understanding of both normal and malignant haematopoiesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Mundlos, S. Cleidocranial dysplasia: clinical and molecular genetics. J. Med. Genet. 36, 177–182 (1999).
Miyoshi, H. et al. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl Acad. Sci. USA 88, 10431–10434 (1991).
Daga, A., Tighe, J. E. & Calabi, F. Leukaemia/Drosophila homology. Nature 356, 484 (1992).
Kagoshima, H. et al. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 9, 338–341 (1993).
Duffy, J. B. & Gergen, J. P. Sex, segments, and the central nervous system: common genetic mechanisms of cell fate determination. Adv. Genet. 31, 1–28 (1994).
Kamachi, Y. et al. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J. Virol. 64, 4808–4819 (1990).
Wang, S. & Speck, N. A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol. Cell. Biol. 12, 89–102 (1992).
Ogawa, E. et al. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila Runt-related DNA binding protein PEBP2α. Virology 194, 314–331 (1993).
Ogawa, E. et al. PEBP2/PEA2 represents a new family of transcription factor homologous to the products of the Drosophila runt and the human AML1 gene. Proc. Natl Acad. Sci. USA 90, 6859–6863 (1993).
Wang, S. et al. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13, 3324–3339 (1993).
Liu, P. et al. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science 261, 1041–1044 (1993).
Huang, G. et al. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteosome-mediated degradation. EMBO J. 20, 723–733 (2001).
Li, L. H. & Gergen, J. P. Differential interactions between Brother proteins and Runt domain proteins in the Drosophila embryo and eye. Development 126, 3313–3322 (1999).
Sakuma, T. et al. Cloning and expression pattern of a novel PEBP2β-binding protein (charged amino acid rich leucine zipper-1[Crl-1]) in the mouse. Mech. Dev. 104, 151–154 (2001).
Okuda, T., van Deursen, J., Hiebert, S. W., Grosveld, G. & Downing, J. R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996).
Wang, Q. et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA 93, 3444–3449 (1996).These two reports characterize the phenotype of Runx1 -knockout mice, and show an absolute requirement for Runx1 for definitive haematopoiesis.
Wang, Q. et al. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87, 697–708 (1996).
Sasaki, K. et al. Absence of fetal liver hematopoiesis in transcriptional co-activator, core binding factor β (Cbfb) deficient mice. Proc. Natl Acad. Sci. USA 93, 12359–12363 (1996).
Giese, K., Kingsley, C., Kirshner, J. R. & Grosschedl, R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA binding and multiple protein–protein interactions. Genes Dev. 9, 995–1008 (1995).
Uchida, H., Zhang, J. & Nimer, S. D. AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol. 158, 2251–2258 (1997).
Wargnier, A. et al. Identification of human granzyme B promoter regulatory elements interacting with activated T-cell-specific proteins: implication of Ikaros and CBF binding sites in promoter activation. Proc. Natl Acad. Sci. USA 92, 6930–6934 (1995).
Takahashi, A. et al. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor (GM-CSF) promoter activity by AML1-related transcription factor, PEBP2. Blood 86, 607–616 (1995).
Brown, S. T. et al. Regulation of the RAG1 promoter by the NF-Y transcription factor. J. Immunol. 158, 5071–5074 (1997).
Armesilla, A. L., Calvo, D. & Vega, M. A. Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J. Biol. Chem. 271, 7781–7787 (1996).
Lutterbach, B. et al. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275, 651–656 (2000).
Palis, J., Robertson, S., Kennedy, M., Wall, C. & Keller, G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084 (1999).
Godin, I., Dieterlen-Lièvre, F. & Cumano, A. Emergence of multipotent hematopoietic cells in the yolk sac and paraaortic splanchnopleura of 8.5 dpc mouse embryos. Proc. Natl Acad. Sci. USA 92, 773–777 (1995).
de Bruijn, M. F. T. R., Speck, N. A., Peeters, M. C. E. & Dzierzak, E. Definitive hematopoietic stem cells first emerge from the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474 (2000).
Niki, M. et al. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl Acad. Sci. USA 94, 5697–5702 (1997).
Cai, Z. et al. Haploinsufficiency of AML1/CBFA2 affects the embryonic generation of mouse hematopoietic stem cells. Immunity 13, 423–431 (2000).
Castilla, L. H. et al. Defects of embryonic hematopoiesis and lethal hemorrhaging in mouse embryos heterozygous for a knocked-in leukemia gene CBFB–MYH11. Cell 87, 687–696 (1996).Shows that the CBFβ–SMMHC fusion protein is a dominant inhibitor of Runx1–CBFβ function during development.
Yokomizo, T. et al. RUNX1/AML1/PEBP2αB is involved in primitive hematopoiesis in addition to definitive hematopoiesis. Blood 96, 283a (2000).
North, T. E. et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563–2575 (1999).This study shows the requirement for Runx1 for the development of haematopoietic clusters in several main arteries during development.
Mukouyama, Y. et al. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta–gonad–mesonephros region. Dev. Biol. 220, 27–36 (2000).
Yokomizo, T. et al. Requirement of Runx1–AML1–PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes Cells 6, 13–23 (2001).
Garcia-Porrero, J. A., Godin, I. E. & Dieterlen-Lièvre, F. Potential intraembryonic hemogenic sites at pre-liver stages in the mouse. Anat. Embryol. 192, 425–435 (1995).
Tavian, M. et al. Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 87, 67–72 (1996).
Jaffredo, T., Gautier, R., Brajeul, V. & Dieterlen-Lièvre, F. Tracing the progeny of the aortic hemangioblast in the avian embryo. Dev. Biol. 224, 204–214 (2000).
North, T. E. et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 (2002).
Lacoud, G. et al. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood (in the press).
Hayashi, K. et al. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 165, 6816–6824 (2000).
Cottles, G. M. et al. Hematological analysis of AML1–PEBP2αB+/− mice; a possible predisposition to myeloid leukemia. Blood 10, 218b (1999).
Ho, C. Y. et al. Linkage of a familial platelet disorder with a propensity to develop myeloid malignancies to human chromosome 21q22.1-22.2. Blood 87, 5218–5224 (1996).
Song, W.-J. et al. Haploinsufficiency of CBFA2 (AML1) causes familial thrombocytopenia with propensity to develop acute myelogenous leukamia (FPD/AML). Nature Genet. 23, 166–175 (1999).Shows, by positional cloning, that an inherited leukaemia syndrome is the consequence of loss of function of RUNX1.
Michaud, J. et al. In vitro analyses of known and novel RUNX1–AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood 99, 1364–1372 (2002).
Osato, M. et al. Biallelic and heterozygous point mutations in the Runt domain of the AML1–PEBP2αB gene associated with myeloblastic leukemias. Blood 93, 1817–1824 (1999).
Preudhomme, C. et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2αB gene in M0 acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood 96, 2862–2869 (2000).
Dal Cin, P. et al. Amplification of AML1 in childhood acute lymphoblastic leukemias. Genes Chromosom. Cancer 30, 407–409 (2001).
Hilgenfeld, E. et al. Spectral karyotyping and fluorescence in situ hybridization detect novel chromosomal aberrations, a recurring involvement of chromosome 21 and amplification of the MYC oncogene in acute myeloid leukaemia M2. Br. J. Haematol. 113, 305–317 (2001).
Mathew, S., Rao, P. H., Dalton, J., Downing, J. R. & Raimondi, S. C. Multicolor spectral karyotyping identifies novel translocations in childhood acute lymphoblastic leukemia. Leukemia 15, 468–472 (2001).
Niini, T., Kanerva, J., Vettenranta, K., Saarinen-Pihkala, U. M. & Knuutila, S. AML1 gene amplification: a novel finding in childhood acute lymphoblastic leukemia. Haematologica 85, 362–366 (2000).
Streubel, B., Valent, P., Lechner, K. & Fonatsch, C. Amplification of the AML1 (CBFA2) gene on ring chromosomes in a patient with acute myeloid leukemia and a constitutional ring chromosome 21. Cancer Genet. Cytogenet. 124, 42–46 (2001).
Stewart, M. et al. Proviral insertions induce the expression of bone-specific isoforms of PEBP2αA (CBFA1): evidence for a new myc collaborating oncogene. Proc. Natl Acad. Sci. USA 94, 8646–8651 (1997).
Blyth, K. et al. Sensitivity to Myc-induced apoptosis is retained in spontaneous and transplanted lymphomas of CD2-mycER mice. Oncogene 19, 773–782 (2000).
Blyth, K. et al. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 20, 295–302 (2001).
Li, J. et al. Leukaemia disease genes: large-scale cloning and pathway predictions. Nature Genet. 23, 348–353 (1999).
Downing, J. R. et al. An AML1–ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the (8;21)(q22;q22) translocation. Blood 81, 2860–2865 (1993).
Erickson, P. et al. Identification of breakpoints in t(8;21) AML and isolation of a fusion transcript with similarity to Drosophila segmentation gene runt. Blood 80, 1825–1831 (1992).
Meyers, S., Downing, J. R. & Hiebert, S. W. Identification of AML1 and the (8;21) translocation protein (AML1–ETO) as sequence-specific DNA-binding proteins: the Runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13, 6336–6345 (1993).
Nucifora, G. et al. Detection of DNA rearrangement in the AML1 and ETO loci and of an AML1–ETO fusion mRNA in patients with t(8;21). Blood 81, 883 (1993).
Golub, T. R. et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 92, 4917–4921 (1995).
Romana, S. P. et al. The t(12;21) of acute lymphoblastic leukemia results in a TEL–AML1 gene fusion. Blood 85, 3662–3670 (1995).
McLean, T. W. et al. TEL–AML1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood 88, 4252–4258 (1996).
Romana, S. P. et al. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood 86, 4263–4269 (1995).
Liang, D. C. et al. High incidence of TEL–AML1 fusion resulting from a cryptic t(12;21) in childhood B-lineage acute lymphoblastic leukemia in Taiwan. Leukemia 10, 991–993 (1996).
Mitani, K. et al. Generation of the AML1–EVI1 fusion gene in the t(3, 21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 13, 504–510 (1994).
Nucifora, G. et al. Consistent intergenic splicing and production of multiple transcripts between AML1 at 21q22 and unrelated genes at 3q26 in (3;21)(q26;q22) translocations. Proc. Natl Acad. Sci. USA 91, 4004–4008 (1994).
Meyers, S., Lenny, N. & Hiebert, S. W. The t(8;21) fusion protein interferes with AML1–1B-dependent transcriptional activation. Mol. Cell. Biol. 15, 1974–1982 (1995).
Hiebert, S. W. et al. The t(12;21) translocation converts AML1B from an activator to a repressor of transcription. Mol. Cell. Biol. 16, 1349–1355 (1996).
Lutterbach, B., Hou, Y., Durst, K. L. & Hiebert, S. W. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc. Natl Acad. Sci. USA 96, 12822–12827 (1999).
Yergeau, D. A. et al. Embryonic lethality and impairment of hematopoiesis in mice heterozygous for an AML1–ETO fusion gene. Nature Genet. 15, 303–306 (1997).
Okuda, T. et al. Expression of a knocked-in AML1–ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood 91, 3134–3143 (1998).These two reports show that the RUNX1–ETO fusion has dominant-negative activity for the native RUNX1 during development.
Shimizu, K. et al. AML1–MTG8 leukemic protein induces the expression of granulocyte colony-stimulating factor (G-CSF) receptor through the up-regulation of CCAAT/enhancer binding protein epsilon. Blood 96, 288–296 (2000).
Rhoades, K. L. et al. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc. Natl Acad. Sci. USA 93, 11895–11900 (1996).
Mulloy, J. C. et al. The AML1–ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 99, 15–23 (2002).
Lutterbach, B. et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18, 7176–7184 (1998).
Gelmetti, V. et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18, 7185–7191 (1998).
Fenrick, R. et al. Both TEL and AML1 contribute repression domains to the t(12;21) fusion protein. Mol. Cell. Biol. 19, 6566–6574 (1999).
Adja, N., Stacy, T., Speck, N. A. & Liu, P. P. The leukemic protein CBFβ–SMMHC sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol. Cell. Biol. 18, 7432–7443 (1998).
Kanno, Y., Kanno, T., Sakakura, C., Bae, S.-C. & Ito, Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2–CBFβ–SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol. 18, 4252–4261 (1998).
Chen, Z. et al. PLZF–RARα fusion proteins generated from the variant t(11;17)(q23;q21) translocation in acute promyelocytic leukemia inhibit ligand-dependent transactivation of wild-type retinoic acid receptors. Proc. Natl Acad. Sci. USA 91, 1178–1182 (1994).
Melnick, A. & Licht, J. D. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93, 3167–3215 (1999).
Melnick, A. et al. AML1–ETO fusion protein is a dominant negative inhibitor of transcriptional repression by the promyelocytic leukemia zinc finger protein. Blood 96, 3939–3947 (2000).
Melnick, A. M. et al. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 20, 2075–2086 (2000).
Minucci, S. et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell 5, 811–820 (2000).
Lin, R. J. & Evans, R. M. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol. Cell 5, 821–830 (2000).
Zhang, J. et al. Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell Biol. 21, 156–163 (2001).
Wang, J., Saunthararajah, Y., Redner, R. L. & Liu, J. M. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1–ETO leukemia cells. Cancer Res. 59, 2766–2769 (1999).
Westendorf, J. J. et al. The t(8;21) fusion product, AML1–ETO, associates with C/EBPα, inhibits C/EBPα-dependent transcription, and blocks granulocytic differentiation. Mol. Cell. Biol. 18, 322–333 (1998).
Wiemels, J. L. et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 354, 1499–1503 (1999).Elegant demonstration, using Guthrie cards, of the prenatal origin of TEL–RUNX1 leukaemia in paediatric ALL, and of the multistep pathogenesis of ALL.
Wiemels, J. L., Ford, A. M., van Wering, E. R., Postma, A. & Greaves, M. Protracted and variable latency of acute lymphoblastic leukemia after TEL–AML1 gene fusion in utero. Blood 94, 1057–1062 (1999).
Ford, A. M. et al. Fetal origins of the TEL–AML1 fusion gene in identical twins with leukemia. Proc. Natl Acad. Sci. USA 95, 4584–4588 (1998).
Downing, J. R. AML1–CBFβ transcription complex: its role in normal hematopoiesis and leukemia. Leukemia 15, 664–665 (2001).
Higuchi, M. et al. Expression of a conditional AML1–ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 1, 63–74 (2002).
Castilla, L. H. et al. Chromosome 16 inversion-generated fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukemia. Nature Genet. 23, 144–146 (1999).These two reports show convincingly that second mutations are required to cause AML that is mediated by the RUNX1–ETO and CBFβ–SMMHC fusion proteins, respectively.
Kundu, M. & Liu, P. P. Function of the inv(16) fusion gene CBFB–MYH11. Curr. Opin. Hematol. 8, 201–205 (2001).
Gilliland, G., Patterson, E. J., Rosenthal, D. S. & Tantravahi, R. Multiple clones with (3;21) translocation in a case of Ph-positive chronic myelogenous leukemia during relapse after allogeneic bone marrow transplantation. Cancer Genet. Cytogenet. 47, 55–60 (1990).
Melo, J. V. The molecular biology of chronic myeloid leukaemia. Leukemia 10, 751–756 (1996).
Kojima, K. et al. Additional translocation (8;21)(q22;q22) in a patient with Philadelphia-positive chronic myelogenous leukaemia in the blastic phase. Br. J. Haematol. 106, 720–722 (1999).
Golub, T. R., Barker, G. F., Lovett, M. & Gilliland, D. G. Fusion of PDGF receptor-β to a novel ETS-like gene, TEL, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77, 307–316 (1994).
Daley, G. Q., Van Etten, R. A. & Baltimore, D. Induction of chronic myelogenous leukemia in mice by the P210BCR–ABL gene of the Philadelphia chromosome. Science 247, 824–830 (1990).
Tomasson, M. H. et al. Fatal myeloproliferation, induced in mice by Tel–Pdgf β r expression, depends on Pdgfβr tyrosines 579/581. J. Clin. Invest. 105, 423–432 (2000).
Nakao, M. et al. Internal tandem duplication of the FLT3 gene found in acute myeloid leukemia. Leukemia 10, 1911–1918 (1996).
Rombouts, W., Blokland, I., Lowenberg, B. & Ploemacher, R. Biological characteristics and prognosis of adult acute myeloid leukemia with internal tandem duplications in the FLT3 gene. Leukemia 14, 675–683 (2000).
Kottaridis, P. D. et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 98, 1752–1759 (2001).
Radich, J. P. et al. N-RAS mutations in adult de novo acute myelogenous leukemia: prevalence and clinical significance. Blood 76, 801–807 (1990).
Neubauer, A. et al. Prognostic importance of mutations in the ras proto-oncogenes in de novo acute myeloid leukemia. Blood 83, 1603–1611 (1994).
Kubo, K. et al. Clonal analysis of multiple point mutations in the N-ras gene in patients with acute myeloid leukemia. Jpn. J. Cancer Res. 84, 379–387 (1993).
Byrd, J. C. et al. Patients with t(8;21)(q22;q22) and acute myeloid leukemia have superior failure-free and overall survival when repetitive cycles of high-dose cytarabine are administered. J. Clin. Oncol. 17, 3767–3775 (1999).
Marcucci, G., Caligiuri, M. A. & Bloomfield, C. D. Molecular and clinical advances in core binding factor primary acute myeloid leukemia: a paradigm for translational research in malignant hematology. Cancer Invest. 18, 768–780 (2000).
Mrozek, K., Heinonen, K. & Bloomfield, C. D. Prognostic value of cytogenetic findings in adults with acute myeloid leukemia. Int. J. Hematol. 72, 261–271 (2000).
Mrozek, K., Heinonen, K. & Bloomfield, C. D. Clinical importance of cytogenetics in acute myeloid leukaemia. Best Pract. Res. Clin. Haematol. 14, 19–47 (2001).
Shurtleff, S. A. et al. TEL–AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia 9, 1985–1989 (1995).
Rubnitz, J. E. et al. TEL gene rearrangement in acute lymphoblastic leukemia: a new genetic marker with prognostic significance. J. Clin. Oncol. 15, 1150–1157 (1997).
Rubnitz, J. E. et al. Case–control study suggests a favorable impact of TEL rearrangement in patients with B-lineage acute lymphoblastic leukemia treated with antimetabolite-based therapy: a Pediatric Oncology Group study. Blood 89, 1143–1146 (1997).
Rubnitz, J. E., Downing, J. R. & Pui, C. H. Significance of the TEL–AML fusion gene in childhood AML. Leukemia 13, 1470–1471 (1999).
Loh, M. L. et al. Incidence of TEL–AML1 fusion in children with relapsed acute lymphoblastic leukemia. Blood 92, 4792–4797 (1998).
Borkhardt, A. et al. Incidence and clinical relevance of TEL–AML1 fusion genes in children with acute lymphoblastic leukemia enrolled in the German and Italian multicenter therapy trials. Associazione Italiana Ematologia Oncologia Pediatrica and the Berlin-Frankfurt-Munster Study Group. Blood 90, 571–577 (1997).
Borkhardt, A., Harbott, J. & Lampert, F. Biology and clinical significance of the TEL–AML1 rearrangement. Curr. Opin. Pediatr. 11, 33–38 (1999).
Harbott, J., Viehmann, S., Borkhardt, A., Henze, G. & Lampert, F. Incidence of TEL–AML1 fusion gene analyzed consecutively in children with acute lymphoblastic leukemia in relapse. Blood 90, 4933 (1997).
Druker, B. J. et al. Activity of a specific inhibitor of the BCR–ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344, 1038–1042 (2001).
Müller, A. M., Medvinsky, A., Strouboulis, J., Grosveld, F. & Dzierzak, E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301 (1994).
Medvinsky, A. & Dzierzak, E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906 (1996).
Choi, K., Kennedy, M., Kazarov, A., Papadimitriou, J. C. & Keller, G. A common precursor for hematopoietic and endothelial cells. Development 125, 725–732 (1998).
Nishikawa, S.-I. et al. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity 8, 761–769 (1998).
Jaffredo, T., Gautier, R., Eichmann, A. & Dieterlen-Lièvre, F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125, 4575–4583 (1998).
Kanno, T. et al. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor-α subunit revealed in the presence of the β-subunit. Mol. Cell. Biol. 18, 2444–2454 (1998).
Gu, T.-L., Goetz, T. L., Graves, B. J. & Speck, N. A. Autoinhibition and partner proteins, CBFβ and Ets-1, modulate DNA binding by CBFα2(AML1). Mol. Cell. Biol. 20, 91–103 (2000).
Zeng, C. et al. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc. Natl Acad. Sci. USA 94, 6746–6751 (1997).
Zeng, C. et al. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl Acad. Sci. USA 95, 1585–1589 (1998).
Ahn, M.-Y. et al. Negative regulation of granulocytic differentiation in the myeloid precursor cell Line 32Dc13 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1–ETO(MTG8). Proc. Natl Acad. Sci. USA 95, 1812–1817 (1998).
Bruhn, L., Munnerlyn, A. & Grosschedl, R. ALY, a context-dependent coactivator of LEF1 and AML1, is required for TCRα enhancer function. Genes Dev. 11, 640–653 (1997).
Petrovick, M. S. et al. Multiple functional domains of AML1: PU. 1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol. 18, 3915–3925 (1998).
Yagi, R., Chen, L.-F., Shigesda, K., Murakami, Y. & Ito, Y. A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 (1999).
Zhang, D.-E. et al. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol. 16, 1231–1240 (1996).
Libermann, T. A. et al. AML1 (CBFα2) cooperates with B cell-specific activating protein (BSAP/PAX5) in activation of the B cell-specific BLK gene promoter. J. Biol. Chem. 274, 24671–24676 (1999).
Mao, S., Frank, R. C., Zhang, J., Miyazaki, Y. & Nimer, S. D. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell. Biol. 19, 3635–3644 (1999).
Kitabayashi, I., Yokoyama, A., Shimizu, K. & Ohki, M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17, 2994–3002 (1998).
Levanon, D. et al. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA 95, 11590–11595 (1998).
Aronson, B. D., Fisher, A. L., Blechman, K., Caudy, M. & Gergen, J. P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol. 17, 5581–5587 (1997).
Chen, L.-F., Ito, K., Murakami, Y. & Ito, Y. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol. Cell. Biol. 18, 4165–4176 (1998).
Miller, J. D., Stacy, T., Liu, P. P. & Speck, N. A. CBFβ, but not CBFβ-SMMHC rescues definitive hematopoiesis in CBFβ-deficient embryonic stem cells. Blood 97, 2248–2256 (2001).
Tang, Y.-Y. et al. Energetic and functional contribution of residues in the core binding factor-β (CBFβ) subunit to heterodimerization with CBFα. J. Biol. Chem. 275, 39579–39588 (2000).
Nagata, T. et al. Immunoglobulin motif DNA-binding and heterodimerization for the PEBP2/CBF Runt-domain. Nature Struct. Biol. 6, 615–619 (1999).
Berardi, M. et al. The Ig fold of the core binding factor α Runt domain is a member of a family of structurally and functionally related Ig fold DNA binding domains. Structure Fold Des. 7, 1247–1256 (1999).
Tahirov, T. H. et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104, 755–767 (2001).
Bravo, J., Li, Z., Speck, N. A. & Warren, A. J. The leukaemia-associated AML1 (Runx1)-CBFβ complex functions as a DNA-induced molecular clamp. Nature Struct. Biol. 8, 371–377 (2001).These two reports provide elegant structural characterization of RUNX1 complexed with its heterodimeric partner CBFβ and DNA, indicating a basis for the allosteric control of RUNX1 by CBFβ and DNA.
Rudolph, M. J. & Gergen, J. P. DNA-binding by Ig-fold proteins. Nature Struct. Biol. 8, 384–386 (2001).
Huang, X., Peng, J. W., Speck, N. A. & Bushweller, J. H. Solution structure of core binding factor-β and map of the CBFα binding site. Nature Struct. Biol. 6, 624–627 (1999).
Goger, M. et al. Molecular insights into PEBP2–CBFβ-SMMHC associated acute leukemia revealed from the three-dimensional structure of PEBP2–CBFβ. Nature Struct. Biol. 6, 620–623 (1999).
Tang, Y.-Y. et al. Biophysical characterization of interactions between the core-binding factor-α and -β subunits and DNA. FEBS Lett. 470, 167–172 (2000).
Langabeer, S. E., Gale, R. E., Rollinson, S. J., Morgan, G. J. & Linch, D. C. Mutations of the AML1 gene in acute myeloid leukemia of the FAB types M0 and M7. Genes Chromosom. Cancer 34, 24–32 (2002).
Imai, Y. et al. Mutations of the AML1 gene in myelodysplastic syndrome and their functional implications in leukemogenesis. Blood 96, 3154–3160 (2000).
Buijs, A. et al. A novel CBFA2 single-nucleotide mutation in familial platelet disorder with propensity to develop myeloid malignancies. Blood 98, 2856–2858 (2001).
Lanza, C. et al. The common TEL–AML1 rearrangement does not represent a frequent event in acute lymphoblastic leukaemia occuring in children with Down syndrome. Leukemia 11, 820–821 (1997).
Takahashi, Y. et al. Prognostic significance of TEL–AML1 fusion transcript in childhood B-precursor acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 20, 190–195 (1998).
Ayigad, S. et al. TEL–AML1 fusion transcript designates a favorable outcome with an intensified protocol in childhood acute lymphoblastic leukemia. Leukemia 13, 481–483 (1999).
Maloney, K. et al. TEL–AML1 fusion identifies a subset of children with standard risk acute lymphoblastic leukemia who have an excellent prognosis when treated with therapy that includes single delayed intensification. Leukemia 13, 1708–1712 (1999).
Hann, I. et al. Determinants of outcome after intensified therapy of childhood lymphoblastic leukaemia: results from Medical Research Council United Kingdom acute lymphoblastic leukaemia XI protocol. Br. J. Haematol. 113, 103–114 (2001).
Hubeek, I. et al. TEL–AML1 fusion is not a prognostic factor in Dutch childhood acute lymphoblastic leukaemia. Br. J. Haematol. 113, 254–255 (2001).
Tsang, K. S. et al. TEL–AML1 rearrangement and the prognostic significance in childhood acute lymphoblastic leukemia in Hong Kong. Am. J. Hematol. 68, 91–98 (2001).
Uckun, F. M. et al. Expression of TEL–AML1 fusion transcripts and response to induction therapy in standard risk acute lymphoblastic leukemia. Leuk. Lymphoma 42, 41–56 (2001).
Seeger, K. et al. TEL–AML1 fusion transcript in relapsed childhood acute lymphoblastic leukemia. Blood 91, 1716–1722 (1998).
Loh, M. L. et al. Incidence of TEL–AML1 fusion in children with relapsed acute lymphoblastic leukemia. Blood 92, 4792–4797 (1998).
Rubnitz, J. E. et al. Low frequency of TEL–AML1 in relapsed acute lymphoblastic leukemia supports a favorable prognosis for this genetic subgroup. Leukemia 13, 19–21 (1999).
Zuna, J. et al. Significantly lower relapse rate for TEL–AML1–positive ALL. Leukemia 13, 1633 (1999).
Tracey, D. W. & Speck, N. A. Potential roles for RUNX1 and its orthologues in determining hematopoietic cell fate. Sem. Cell Dev. Biol. 11, 337–342 (2000).
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Cancer.gov
LocusLink
Medscape DrugInfo
OMIM
Glossary
- POLYOMAVIRUS
-
A papovavirus of rodents that is associated with various kinds of tumour.
- ENHANCER
-
A regulatory sequence in eukaryotic DNA that might be located at a great distance, either upstream or downstream, from the transcriptional start site of the gene that it controls.
- PHENOCOPY
-
A genotypic variant that resembles the normal expression of a genotype other than its own.
- PARA-AORTIC SPLANCHNOPLEURE
-
(PAS). A layer of tissue near the aorta that consists of the inner of the two layers into which the lateral plate of the mesoderm splits in the embryo of a crainate vertebrate, together with the endoderm internal to it, and which forms most of the walls and substance of the visceral organs.
- VITELLINE ARTERY
-
Also known as the omphalomesenteric artery. An artery that arises in a vertebrate embryo from the aorta or one of the aortic trunks of the embryo, and is distributed by numerous branches over the yolk sac.
- UMBILICAL ARTERY
-
Either of a pair of arteries that arise from the hypogastric arteries of the mammalian fetus and pass through the umbilical cord to the placenta, to which they carry deoxygenated blood from the fetus.
- AORTA/GONAD/MESONEPHROS
-
(AGM). The site of the developing aorta, gonads and renal organs; a site of early haematopoietic development.
- THROMBOCYTOPENIA
-
A persistent decrease in the number of platelets.
- FLUORESCENCE IN SITU HYBRIDIZATION
-
(FISH). A generalized technique for visualizing cellular components, including chromosomes, by treating cells with a fluorescently labelled agent that binds to the component of interest, and observing the cells with fluorescence microscopy. In this context, fluorescent DNA probes for the RUNX1 locus might be used to detect the t(12;21)(p13;q22) TEL–RUNX1 translocation, as well as amplification of the locus.
- BLAST CRISIS
-
Chronic myelogenous leukaemia (CML) is a chronic myeloproliferative syndrome that progresses to acute leukaemia with the acquisition of additional mutations. Acute leukaemia in this context is referred to as 'blast crisis', and is almost invariably a fatal complication of CML.
- GUTHRIE CARD
-
A card onto which a small sample of blood is collected in all neonates of developed countries to screen for metabolic and inherited disease. Originally used for the 'Guthrie test' for phenylketonuria. The cards contain small amounts of DNA from leukocytes that are present in the blood sample, and can be screened for mutations by polymerase-chain-reaction-based strategies.
Rights and permissions
About this article
Cite this article
Speck, N., Gilliland, D. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer 2, 502–513 (2002). https://doi.org/10.1038/nrc840
Issue Date:
DOI: https://doi.org/10.1038/nrc840
This article is cited by
-
m6A genotypes and prognostic signature for assessing the prognosis of patients with acute myeloid leukemia
BMC Medical Genomics (2023)
-
RUNX1::ETO translocations must precede CSF3R mutations to promote acute myeloid leukemia development
Leukemia (2023)
-
CAG (cytarabine, aclarubicin and granulocyte colony-stimulating factor) regimen for core binding factor acute myeloid leukaemia with measurable residual disease
Annals of Hematology (2023)
-
N6-methyladenosine in myeloid cells: a novel regulatory factor for inflammation-related diseases
Journal of Physiology and Biochemistry (2023)
-
Neoantigen-specific TCR-T cell-based immunotherapy for acute myeloid leukemia
Experimental Hematology & Oncology (2022)