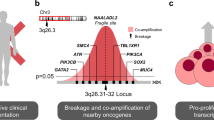
Abstract
Ever since initial suggestions that instability at common fragile sites (CFSs) could be responsible for chromosome rearrangements in cancers, CFSs and associated genes have been the subject of numerous studies, leading to questions and controversies about their role and importance in cancer. It is now clear that CFSs are not frequently involved in translocations or other cancer-associated recurrent gross chromosome rearrangements. However, recent studies have provided new insights into the mechanisms of CFS instability, their effect on genome instability, and their role in generating focal copy number alterations that affect the genomic landscape of many cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Glover, T. W., Berger, C., Coyle, J. & Echo, B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67, 136–142 (1984).
Yunis, J. J. Fragile sites and predisposition to leukemia and lymphoma. Cancer Genet. Cytogenet. 12, 85–88 (1984).
de Braekeleer, M. Fragile sites and chromosomal structural rearrangements in human leukemia and cancer. Anticancer Res. 7, 417–422 (1987).
Yunis, J. J. Fragile sites, mutagens and genomic rearrangements in cancer. Basic Life Sci. 43, 11–21 (1988).
Le Beau, M. M. Chromosomal fragile sites and cancer-specific rearrangements. Blood 67, 849–858 (1986).
De Braekeleer, M., Smith, B. & Lin, C. C. Fragile sites and structural rearrangements in cancer. Hum. Genet. 69, 112–116 (1985).
Barlow, J. H. et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013).
Huebner, K., Hadaczek, P., Siprashvili, Z., Druck, T. & Croce, C. M. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Biochim. Biophys. Acta 1332, M65–M70 (1997).
Ohta, M. et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 84, 587–597 (1996).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Yan, Z. A., Li, X. Z. & Zhou, X. T. The effect of hydroxyurea on the expression of the common fragile site at 3p14. J. Med. Genet. 24, 593–596 (1987).
[No authors listed.] An international system for human cytogenetic nomenclature — high-resolution banding (1981). ISCN (1981). Report of the Standing Committee on Human Cytogenetic Nomenclature. Cytogenet. Cell Genet. 31, 5–23 (1981).
Mariani, T. Fragile sites and statistics. Hum. Genet. 81, 319–322 (1989).
Glover, T. W. & Stein, C. K. Induction of sister chromatid exchanges at common fragile sites. Am. J. Hum. Genet. 41, 882–890 (1987).
Wenger, S. L. Chemical induction of sister chromatid exchange at fragile sites. Cancer Genet. Cytogenet. 85, 72–74 (1995).
Glover, T. W. & Stein, C. K. Chromosome breakage and recombination at fragile sites. Am. J. Hum. Genet. 43, 265–273 (1988).
Wang, N. D., Testa, J. R. & Smith, D. I. Determination of the specificity of aphidicolin-induced breakage of the human 3p14.2 fragile site. Genomics 17, 341–347 (1993).
Rassool, F. V. et al. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc. Natl Acad. Sci. USA 88, 6657–6661 (1991).
Durkin, S. G. & Glover, T. W. Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 (2007).
Sarni, D. & Kerem, B. The complex nature of fragile site plasticity and its importance in cancer. Curr. Opin. Cell Biol. 40, 131–136 (2016).
Arlt, M. F., Casper, A. M. & Glover, T. W. Common fragile sites. Cytogenet. Genome Res. 100, 92–100 (2003).
Le Beau, M. M. et al. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 7, 755–761 (1998).
Wilke, C. M. et al. Multicolor FISH mapping of YAC clones in 3p14 and identification of a YAC spanning both FRA3B and the t(3;8) associated with hereditary renal cell carcinoma. Genomics 22, 319–326 (1994).
Paradee, W. et al. Precise localization of aphidicolin-induced breakpoints on the short arm of human chromosome 3. Genomics 27, 358–361 (1995).
Mangelsdorf, M. et al. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 60, 1683–1689 (2000).
Bednarek, A. K. et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 60, 2140–2145 (2000).
Smith, D. I., Zhu, Y., McAvoy, S. & Kuhn, R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 232, 48–57 (2006).
Gao, G. & Smith, D. I. Very large common fragile site genes and their potential role in cancer development. Cell. Mol. Life Sci. 71, 4601–4615 (2014).
Wilson, T. E. et al. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 25, 189–200 (2015).
Murano, I., Kuwano, A. & Kajii, T. Cell type-dependent difference in the distribution and frequency of aphidicolin-induced fragile sites: T and B lymphocytes and bone marrow cells. Hum. Genet. 84, 71–74 (1989).
Murano, I., Kuwano, A. & Kajii, T. Fibroblast-specific common fragile sites induced by aphidicolin. Hum. Genet. 83, 45–48 (1989).
Kuwano, A., Murano, I. & Kajii, T. Cell type-dependent difference in the distribution and frequency of excess thymidine-induced common fragile sites: T lymphocytes and skin fibroblasts. Hum. Genet. 84, 527–531 (1990).
Le Tallec, B. et al. Molecular profiling of common fragile sites in human fibroblasts. Nat. Struct. Mol. Biol. 18, 1421–1423 (2011).
Le Tallec, B. et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 4, 420–428 (2013).
Hosseini, S. A. et al. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer 52, 1017–1029 (2013).
Glover, T. W. in Genetic Instabilities and Neurological Diseases (eds Wells, R. D. & Warren, S. T.) 75–83 (Academic Press, 1998).
Palakodeti, A. et al. Impaired replication dynamics at the FRA3B common fragile site. Hum. Mol. Genet. 19, 99–110 (2010).
Glover, T. W., Arlt, M. F., Casper, A. M. & Durkin, S. G. Mechanisms of common fragile site instability. Hum. Mol. Genet. 14 (Suppl. 2), R197–R205 (2005).
Boldog, F. et al. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum. Mol. Genet. 6, 193–203 (1997).
Ried, K. et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 9, 1651–1663 (2000).
Arlt, M. F., Miller, D. E., Beer, D. G. & Glover, T. W. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer 33, 82–92 (2002).
Zlotorynski, E. et al. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 23, 7143–7151 (2003).
Mishmar, D. et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl Acad. Sci. USA 95, 8141–8146 (1998).
Mirkin, E. V. & Mirkin, S. M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71, 13–35 (2007).
Zhang, H. & Freudenreich, C. H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell 27, 367–379 (2007).
Durkin, S. G. et al. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl Acad. Sci. USA 105, 246–251 (2008).
Casper, A. M., Nghiem, P., Arlt, M. F. & Glover, T. W. ATR regulates fragile site stability. Cell 111, 779–789 (2002).
Durkin, S. G., Arlt, M. F., Howlett, N. G. & Glover, T. W. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 25, 4381–4388 (2006).
Zhu, M. & Weiss, R. S. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol. Biol. Cell 18, 1044–1055 (2007).
Focarelli, M. L. et al. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer 48, 1083–1090 (2009).
Musio, A. et al. SMC1 involvement in fragile site expression. Hum. Mol. Genet. 14, 525–533 (2005).
Bhat, A., Andersen, P. L., Qin, Z. & Xiao, W. Rev3, the catalytic subunit of Polzeta, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 41, 2328–2339 (2013).
Madireddy, A. et al. FANCD2 facilitates replication through common fragile sites. Mol. Cell 64, 388–404 (2016).
Bhowmick, R., Minocherhomji, S. & Hickson, I. D. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol. Cell 64, 1117–1126 (2016).
Mason, J. M. et al. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum. Mol. Genet. 22, 4901–4913 (2013).
Ying, S. et al. MUS81 promotes common fragile site expression. Nat. Cell Biol. 15, 1001–1007 (2013).
Naim, V., Wilhelm, T., Debatisse, M. & Rosselli, F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15, 1008–1015 (2013).
Minocherhomji, S. et al. Replication stress activates DNA repair synthesis in mitosis. Nature 528, 286–290 (2015).
Bergoglio, V. et al. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 201, 395–408 (2013).
Walsh, E., Wang, X., Lee, M. Y. & Eckert, K. A. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J. Mol. Biol. 425, 232–243 (2013).
Pirzio, L. M., Pichierri, P., Bignami, M. & Franchitto, A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 180, 305–314 (2008).
Fundia, A., Gorla, N. & Larripa, I. Non-random distribution of spontaneous chromosome aberrations in two Bloom Syndrome patients. Hereditas 122, 239–243 (1995).
Howlett, N. G., Taniguchi, T., Durkin, S. G., D'Andrea, A. D. & Glover, T. W. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 14, 693–701 (2005).
Arlt, M. F. et al. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 24, 6701–6709 (2004).
Schwartz, M. et al. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 19, 2715–2726 (2005).
Letessier, A. et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 (2011).
Miotto, B., Ji, Z. & Struhl, K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl Acad. Sci. USA 113, E4810–E4819 (2016).
Sotiriou, S. K. et al. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell 64, 1127–1134 (2016).
Baumann, C., Korner, R., Hofmann, K. & Nigg, E. A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 (2007).
Chan, K. L. & Hickson, I. D. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 22, 906–912 (2011).
Biebricher, A. et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell 51, 691–701 (2013).
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015).
Zhang, F., Gu, W., Hurles, M. E. & Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481 (2009).
Lee, C. & Scherer, S. W. The clinical context of copy number variation in the human genome. Expert Rev. Mol. Med. 12, e8 (2010).
McConnell, M. J. et al. Mosaic copy number variation in human neurons. Science 342, 632–637 (2013).
Campbell, I. M. et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am. J. Hum. Genet. 95, 173–182 (2014).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Arlt, M. F. et al. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 84, 339–350 (2009).
Arlt, M. F., Ozdemir, A. C., Birkeland, S. R., Wilson, T. E. & Glover, T. W. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl Acad. Sci. USA 108, 17360–17365 (2011).
Arlt, M. F., Rajendran, S., Birkeland, S. R., Wilson, T. E. & Glover, T. W. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ. Mol. Mutagen. 55, 103–113 (2014).
Arlt, M. F., Rajendran, S., Birkeland, S. R., Wilson, T. E. & Glover, T. W. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 8, e1002981 (2012).
Liu, P., Carvalho, C. M., Hastings, P. J. & Lupski, J. R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22, 211–220 (2012).
Helmrich, A., Ballarino, M. & Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011).
Newman, T. J., Mamun, M. A., Nieduszynski, C. A. & Blow, J. J. Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts. Nucleic Acids Res. 41, 9705–9718 (2013).
Ozeri-Galai, E. et al. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell 43, 122–131 (2011).
Snyder, M., Sapolsky, R. J. & Davis, R. W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 2184–2194 (1988).
Looke, M. et al. Relicensing of transcriptionally inactivated replication origins in budding yeast. J. Biol. Chem. 285, 40004–40011 (2010).
Wei, P. C. et al. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164, 644–655 (2016).
Wilke, C. M. et al. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 5, 187–195 (1996).
Gao, G. et al. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer 56, 59–74 (2017).
Kraus, I. et al. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 68, 2514–2522 (2008).
Thorland, E. C. et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 60, 5916–5921 (2000).
Wentzensen, N., Vinokurova, S. & von Knebel Doeberitz, M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 64, 3878–3884 (2004).
Matovina, M., Sabol, I., Grubisic, G., Gasperov, N. M. & Grce, M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol. Oncol. 113, 120–127 (2009).
Walline, H. M. et al. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head Neck 39, 840–852 (2017).
Jang, M. K., Shen, K. & McBride, A. A. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 10, e1004117 (2014).
Hu, Z. et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 47, 158–163 (2015).
Doolittle-Hall, J. M., Cunningham Glasspoole, D. L., Seaman, W. T. & Webster-Cyriaque, J. Meta-analysis of DNA tumor-viral integration site selection indicates a role for repeats, gene expression and epigenetics. Cancers (Basel) 7, 2217–2235 (2015).
Finnis, M. et al. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum. Mol. Genet. 14, 1341–1349 (2005).
Denison, S. R., Callahan, G., Becker, N. A., Phillips, L. A. & Smith, D. I. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer 38, 40–52 (2003).
Callahan, G., Denison, S. R., Phillips, L. A., Shridhar, V. & Smith, D. I. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene 22, 590–601 (2003).
Huang, H., Qian, C., Jenkins, R. B. & Smith, D. I. Fish mapping of YAC clones at human chromosomal band 7q31.2: identification of YACS spanning FRA7G within the common region of LOH in breast and prostate cancer. Genes Chromosomes Cancer 21, 152–159 (1998).
Lai, L. A. et al. Deletion at fragile sites is a common and early event in Barrett's esophagus. Mol. Cancer Res. 8, 1084–1094 (2010).
Michael, D., Beer, D. G., Wilke, C. W., Miller, D. E. & Glover, T. W. Frequent deletions of FHIT and FRA3B in Barrett's metaplasia and esophageal adenocarcinomas. Oncogene 15, 1653–1659 (1997).
Gu, J. et al. Genome-wide catalogue of chromosomal aberrations in barrett's esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev. Res. (Phila.) 3, 1176–1186 (2010).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Rajaram, M. et al. Two distinct categories of focal deletions in cancer genomes. PLoS ONE 8, e66264 (2013).
Zhao, M., Kim, P., Mitra, R., Zhao, J. & Zhao, Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 44, D1023–D1031 (2016).
Romagosa, C. et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30, 2087–2097 (2011).
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Halazonetis, T. D., Gorgoulis, V. G. & Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008).
Georgakilas, A. G. et al. Are common fragile sites merely structural domains or highly organized “functional” units susceptible to oncogenic stress? Cell. Mol. Life Sci. 71, 4519–4544 (2014).
Dereli-Oz, A., Versini, G. & Halazonetis, T. D. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol. Oncol. 5, 308–314 (2011).
Gaillard, H., Garcia-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Neelsen, K. J., Zanini, I. M., Herrador, R. & Lopes, M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200, 699–708 (2013).
Teixeira, L. K. et al. Cyclin E deregulation promotes loss of specific genomic regions. Curr. Biol. 25, 1327–1333 (2015).
Kotsantis, P. et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 7, 13087 (2016).
Tsantoulis, P. K. et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene 27, 3256–3264 (2008).
Miron, K., Golan-Lev, T., Dvir, R., Ben-David, E. & Kerem, B. Oncogenes create a unique landscape of fragile sites. Nat. Commun. 6, 7094 (2015).
Karras, J. R., Schrock, M. S., Batar, B. & Huebner, K. Fragile genes that are frequently altered in cancer: players not passengers. Cytogenet. Genome Res. 150, 208–216 (2017).
Schrock, M. S. & Huebner, K. WWOX: a fragile tumor suppressor. Exp. Biol. Med. (Maywood) 240, 296–304 (2015).
Guler, G. et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol. Int. 55, 471–478 (2005).
Sozzi, G. et al. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 58, 5032–5037 (1998).
Wu, X., Wu, G., Yao, X., Hou, G. & Jiang, F. The clinicopathological significance and ethnic difference of FHIT hypermethylation in non-small-cell lung carcinoma: a meta-analysis and literature review. Drug Des. Devel. Ther. 10, 699–709 (2016).
Kandimalla, R., van Tilborg, A. A. & Zwarthoff, E. C. DNA methylation-based biomarkers in bladder cancer. Nat. Rev. Urol. 10, 327–335 (2013).
Baryla, I., Styczen-Binkowska, E. & Bednarek, A. K. Alteration of WWOX in human cancer: a clinical view. Exp. Biol. Med. (Maywood) 240, 305–314 (2015).
Gao, G. & Smith, D. I. WWOX, large common fragile site genes, and cancer. Exp. Biol. Med. (Maywood) 240, 285–295 (2015).
Saldivar, J. C. et al. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet. 8, e1003077 (2012).
Abu-Odeh, M., Hereema, N. A. & Aqeilan, R. I. WWOX modulates the ATR-mediated DNA damage checkpoint response. Oncotarget 7, 4344–4355 (2016).
Drusco, A. et al. Common fragile site tumor suppressor genes and corresponding mouse models of cancer. J. Biomed. Biotechnol. 2011, 984505 (2011).
Aqeilan, R. I. et al. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 67, 5606–5610 (2007).
Zanesi, N. et al. The tumor spectrum in FHIT-deficient mice. Proc. Natl Acad. Sci. USA 98, 10250–10255 (2001).
Aqeilan, R. I. et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl Acad. Sci. USA 104, 3949–3954 (2007).
Gong, Y. et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat. Genet. 46, 588–594 (2014).
Le Beau, M. M. et al. An FHIT tumor suppressor gene? Genes Chromosomes Cancer 21, 281–289 (1998).
Nagamani, S. C. et al. Detection of copy-number variation in AUTS2 gene by targeted exonic array CGH in patients with developmental delay and autistic spectrum disorders. Eur. J. Hum. Genet. 21, 343–346 (2013).
Swaminathan, S. et al. Analysis of copy number variation in Alzheimer's disease: the NIALOAD/ NCRAD Family Study. Curr. Alzheimer Res. 9, 801–814 (2012).
Gregor, A. et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med. Genet. 12, 106 (2011).
Mikhail, F. M. et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am. J. Med. Genet. A 155A, 2386–2396 (2011).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Broad Institute TCGA Genome Data Analysis Center. SNP6 copy number analysis (GISTIC2). Broad Institute https://doi.org/10.7908/C1P84B9Q (2016).
Arlt, M. F. & Glover, T. W. Inhibition of topoisomerase I prevents chromosome breakage at common fragile sites. DNA Repair (Amst.) 9, 678–689 (2010).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
Casper, A. M., Durkin, S. G., Arlt, M. F. & Glover, T. W. Chromosomal instability at common fragile sites in Seckel syndrome. Am. J. Hum. Genet. 75, 654–660 (2004).
Ozeri-Galai, E., Schwartz, M., Rahat, A. & Kerem, B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene 27, 2109–2117 (2008).
Schwartz, M. et al. Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS ONE 4, e4516 (2009).
Acknowledgements
The authors would like to thank J. Moran for critical reading this manuscript and R. Beroukhim for sharing early versions of the The Cancer Genome Atlas (TCGA) copy number variant (CNV) data analyses. This work was supported in part by grants ES020875 from the US National Institute of Environmental Health Sciences and CA 200731 from the US National Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Glover, T., Wilson, T. & Arlt, M. Fragile sites in cancer: more than meets the eye. Nat Rev Cancer 17, 489–501 (2017). https://doi.org/10.1038/nrc.2017.52
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc.2017.52
This article is cited by
-
Chronic replication stress invokes mitochondria dysfunction via impaired parkin activity
Scientific Reports (2024)
-
Genome maintenance meets mechanobiology
Chromosoma (2024)
-
Comprehensive analysis of neoantigens derived from structural variation across whole genomes from 2528 tumors
Genome Biology (2023)
-
Deterministic evolution and stringent selection during preneoplasia
Nature (2023)
-
Integrative analysis reveals early epigenetic alterations in high-grade serous ovarian carcinomas
Experimental & Molecular Medicine (2023)