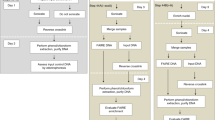
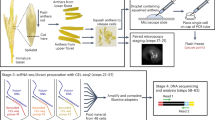
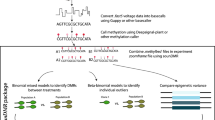
Abstract
The early endosperm tissue of dicot species is very difficult to isolate by manual dissection. This protocol details how to apply the INTACT (isolation of nuclei tagged in specific cell types) system for isolating early endosperm nuclei of Arabidopsis at high purity and how to generate parental-specific epigenome profiles. As a Protocol Extension, this article describes an adaptation of an existing Nature Protocol that details the use of the INTACT method for purification of root nuclei. We address how to obtain the INTACT lines, generate the starting material and purify the nuclei. We describe a method that allows purity assessment, which has not been previously addressed. The purified nuclei can be used for ChIP and DNA bisulfite treatment followed by next-generation sequencing (seq) to study histone modifications and DNA methylation profiles, respectively. By using two different Arabidopsis accessions as parents that differ by a large number of single-nucleotide polymorphisms (SNPs), we were able to distinguish the parental origin of epigenetic modifications. Our protocol describes the only working method to our knowledge for generating parental-specific epigenome profiles of the early Arabidopsis endosperm. The complete protocol, from silique collection to finished libraries, can be completed in 2 d for bisulfite-seq (BS-seq) and 3 to 4 d for ChIP-seq experiments.
This protocol is an extension to: Nat. Protoc. 6, 56–68 (2011); doi:10.1038/nprot.2010.175; published online 16 December 2010
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dresselhaus, T., Sprunck, S. & Wessel, G.M. Fertilization mechanisms in flowering plants. Curr. Biol. 26, R125–R139 (2016).
Deal, R.B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 (2010).
Moreno-Romero, J., Jiang, H., Santos-Gonzalez, J. & Kohler, C. Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35, 1298–1311 (2016).
Deal, R.B. & Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana . Nat. Protoc. 6, 56–68 (2011).
Villar, C.B.R., Erilova, A., Makarevich, G., Trosch, R. & Kohler, C. Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol. Plant 2, 654–660 (2009).
Kohler, C. et al. The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553 (2003).
Lu, P. et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22, 508–18 (2012).
Hsieh, T.F. et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108, 1755–62 (2011).
Weinhofer, I., Hehenberger, E., Roszak, P., Hennig, L. & Kohler, C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6 (2010).
Schatlowski, N. et al. Hypomethylated pollen bypasses the interploidy hybridization barrier in. Plant Cell 26, 3556–68 (2014).
Hsieh, T.F. et al. Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–4 (2009).
Rea, M. et al. Determination of DNA methylation of imprinted genes in Arabidopsis endosperm. J. Vis. Exp. http://dx.doi.org/10.3791/2327 (2011).
Gehring, M., Bubb, K.L. & Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451 (2009).
Le, B.H. et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107, 8063–70 (2010).
Weinhofer, I. & Kohler, C. Endosperm-specific chromatin profiling by fluorescence-activated nuclei sorting and ChIP-on-chip. Methods Mol. Biol. 1112, 105–115 (2014).
Gehring, M., Missirian, V. & Henikoff, S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6, e23687 (2011).
Pignatta, D. et al. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 3, e03198 (2014).
Marques-Bueno, M.M. et al. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in. Plant J. 85, 320–33 (2016).
Clough, S.J. & Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743 (1998).
Przybyla, D. et al. Enzymatic, but not non-enzymatic, O-1(2)-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana . Plant J. 54, 236–248 (2008).
Goto, K. & Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene Pistillata. Genes Dev. 8, 1548–1560 (1994).
Haring, M. et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3, 11 (2007).
Kaufmann, K. et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5, 457–472 (2010).
Li, W. et al. A robust chromatin immunoprecipitation protocol for studying transcription factor-DNA interactions and histone modifications in wood-forming tissue. Nat. Protoc. 9, 2180–2193 (2014).
Saleh, A., Alvarez-Venegas, R. & Avramova, Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025 (2008).
Yamaguchi, N. et al. Protocols: chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12, e0170 (2014).
Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics Chapter 11 Unit 11 7 (2010).
Cheadle, C., Vawter, M.P., Freed, W.J. & Becker, K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5, 73–81 (2003).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Wang, D. & Deal, R.B. Epigenome profiling of specific plant cell types using a streamlined INTACT protocol and ChIP-seq. Methods Mol. Biol. 1284, 3–25 (2015).
Acknowledgements
This research was supported by a European Research Council Starting Independent Researcher grant (to C.K.), a grant from the Swedish Science Foundation (to C.K.), a grant from the Knut and Alice Wallenberg Foundation (to C.K.) and a grant from the Royal Physiographic Society in Lund (to J.M.-R.). We thank A. Ortiz Herrera for recording and editing the video.
Author information
Authors and Affiliations
Contributions
J.M.-R. and C.K. designed the research; J.M.-R. performed the experiments; J.M.-R., J.S.-G. and C.K. analyzed the data; L.H. developed the formula; and J.M.-R. and C.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Binding assay, yield and purity of INTACT purified crosslinked siliques
Comparison of INTACT purified nuclei from cross-linked and non-crosslinked siliques.
a. Bead-binding performed in nuclei extracted from cross-linked siliques (first two panels). Siliques that were either split open or left intact were cross-linked and efficient bead-binding was observed of nuclei extracted from this tissue. Nuclei from non-crosslinked siliques (last two panels) were binding to beads with similar efficiency.
b. (first and second columns) Amount of DNA recovered after purification (pg of DNA and nuclei number considering that an Arabidopsis 3n endosperm nuclei corresponds to 0.45 pg of DNA) and (last column) percentage of Ler left from a 43% (wt/wt) spike-in starting material. The starting material was 200 mg of INT siliques and 150 mg of Ler siliques.
Supplementary Figure 2 Ploidy analysis of the non-purified control sample (Step 17).
a. Ploidy profile of nuclei from adult leaves extracted by chopping ( Box 2 ). 2n, 4n and 8n peaks can be distinguished.
b. Ploidy profile of nuclei extracted from siliques at 4 DAP as described in the main procedure (Steps 1-17). Nuclei from maternal diploid tissue (2n, 4n and 8n) and endosperm nuclei (3n and 6n) can be detected as peaks. Quantifying this profile estimates about 16% of endosperm nuclei.
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1 and 2, Supplementary Tables 1–3, and Supplementary Notes 1–3. (PDF 2908 kb)
Supplementary Data 1
bedGraph file used in Figure 5. (ZIP 24523 kb)
Supplementary Data 2
bedGraph file used in Figure 5. (ZIP 23040 kb)
Supplementary Data 3
bedGraph file used in Figure 5. (ZIP 21602 kb)
Supplementary Data 4
bedGraph file used in Figure 5. (ZIP 24213 kb)
Supplementary Data 5
bedGraph file used in Figure 5. (ZIP 23189 kb)
Supplementary Data 6
bedGraph file used in Figure 5. (ZIP 21828 kb)
Supplementary Data 7
bedGraph file used in Figure 5. (ZIP 17288 kb)
Supplementary Data 8
bedGraph file used in Figure 5. (ZIP 8412 kb)
Supplementary Data 9
bedGraph file used in Figure 5. (ZIP 7858 kb)
Supplementary Data 10
bedGraph file used in Figure 5. (ZIP 8028 kb)
Supplementary Data 11
bedGraph file used in Figure 5. (ZIP 8114 kb)
Supplementary Data 12
Archive containing custom scripts used in Figure 5 (script names and short descriptions listed in Supplementary Table 3). (ZIP 38056 kb)
Rights and permissions
About this article
Cite this article
Moreno-Romero, J., Santos-González, J., Hennig, L. et al. Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat Protoc 12, 238–254 (2017). https://doi.org/10.1038/nprot.2016.167
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.167
This article is cited by
-
The eINTACT method for studying nuclear changes in host plant cells targeted by bacterial effectors in native infection contexts
Nature Protocols (2023)
-
Time series single-cell transcriptional atlases reveal cell fate differentiation driven by light in Arabidopsis seedlings
Nature Plants (2023)
-
FlsnRNA-seq: protoplasting-free full-length single-nucleus RNA profiling in plants
Genome Biology (2021)
-
LEAFY COTYLEDON1 expression in the endosperm enables embryo maturation in Arabidopsis
Nature Communications (2021)
-
Epigenetic signatures associated with imprinted paternally expressed genes in the Arabidopsis endosperm
Genome Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.