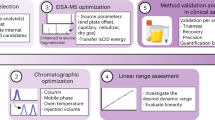
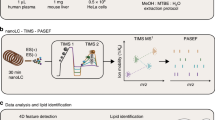
Abstract
Metabolomic and lipidomic studies measure and discover metabolic and lipid profiles in biological samples, enabling a better understanding of the metabolism of specific biological phenotypes. Accurate biological interpretations require high analytical reproducibility and sensitivity, and standardized and transparent data processing. Here we describe a complete workflow for nanoelectrospray ionization (nESI) direct-infusion mass spectrometry (DIMS) metabolomics and lipidomics. After metabolite and lipid extraction from tissues and biofluids, samples are directly infused into a high-resolution mass spectrometer (e.g., Orbitrap) using a chip-based nESI sample delivery system. nESI functions to minimize ionization suppression or enhancement effects as compared with standard electrospray ionization (ESI). Our analytical technique—named spectral stitching—measures data as several overlapping mass-to-charge (m/z) windows that are subsequently 'stitched' together, creating a complete mass spectrum. This considerably increases the dynamic range and detection sensitivity—about a fivefold increase in peak detection—as compared with the collection of DIMS data as a single wide mass-to-charge (m/z ratio) window. Data processing, statistical analysis and metabolite annotation are executed as a workflow within the user-friendly, transparent and freely available Galaxy platform (galaxyproject.org). Generated data have high mass accuracy that enables molecular formulae peak annotations. The workflow is compatible with any sample-extraction method; in this protocol, the examples are extracted using a biphasic method, with methanol, chloroform and water as the solvents. The complete workflow is reproducible, rapid and automated, which enables cost-effective analysis of >10,000 samples per year, making it ideal for high-throughput metabolomics and lipidomics screening—e.g., for clinical phenotyping, drug screening and toxicity testing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
09 February 2017
In the version of this article initially published online, the page numbers were assigned incorrectly as 255–273. The correct page numbers are 310–328. The error has been corrected in the print, PDF and HTML versions of this article.
References
Fiehn, O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Nicholson, J.K., Connelly, J., Lindon, J.C. & Holmes, E. Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 1, 153–161 (2002).
Han, X.L. & Gross, R.W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24, 367–412 (2005).
Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4, 594–610 (2005).
Quehenberger, O. et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51, 3299–3305 (2010).
Sreekumar, A. et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 (2009).
Southam, A.D. et al. Drug redeployment to kill leukemia and lymphoma cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids. Cancer Res. 75, 2530–2540 (2015).
Allen, J. et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003).
Kamphorst, J.J. et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015).
Taylor, N.S. et al. A new approach to toxicity testing in Daphnia magna: application of high throughput FT-ICR mass spectrometry metabolomics. Metabolomics 5, 44–58 (2009).
Southam, A.D. et al. Metabolomics reveals target and off-target toxicities of a model organophosphate pesticide to roach (Rutilus rutilus): implications for biomonitoring. Environ. Sci. Tech. 45, 3759–3767 (2011).
Gruening, N.-M. et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14, 415–427 (2011).
Li, X., Gianoulis, T.A., Yip, K.Y., Gerstein, M. & Snyder, M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell 143, 639–650 (2010).
Kaelin, W.G. Jr. & McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013).
Dunn, W.B., Bailey, N.J.C. & Johnson, H.E. Measuring the metabolome: current analytical technologies. Analyst 130, 606–625 (2005).
Want, E.J., Cravatt, B.F. & Siuzdak, G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem 6, 1941–1951 (2005).
Jonsson, P. et al. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 77, 5635–5642 (2005).
Theodoridis, G., Gika, H.G. & Wilson, I.D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trac-Trends Anal. Chem. 27, 251–260 (2008).
Ramautar, R., Somsen, G.W. & de Jong, G.J. CE-MS for metabolomics: developments and applications in the period 2010-2012. Electrophoresis 34, 86–98 (2013).
Koulman, A. et al. High-resolution extracted ion chromatography, a new tool for metabolomics and lipidomics using a second-generation orbitrap mass spectrometer. Rapid Commun. Mass Spectrom. 23, 1411–1418 (2009).
Castrillo, J.I., Hayes, A., Mohammed, S., Gaskell, S.J. & Oliver, S.G. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry 62, 929–937 (2003).
Beckmann, M., Parker, D., Enot, D.P., Duval, E. & Draper, J. High-throughput, nontargeted metabolite fingerprinting using nominal mass flow injection electrospray mass spectrometry. Nat. Protoc. 3, 486–504 (2008).
Southam, A.D., Payne, T.G., Cooper, H.J., Arvanitis, T.N. & Viant, M.R. Dynamic range and mass accuracy of wide-scan direct infusion nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry-based metabolomics increased by the spectral stitching method. Anal. Chem. 79, 4595–4602 (2007).
Weber, R.J.M., Southam, A.D., Sommer, U. & Viant, M.R. Characterization of isotopic abundance measurements in high resolution FT-ICR and Orbitrap mass spectra for improved confidence of metabolite identification. Anal. Chem. 83, 3737–3743 (2011).
Hop, C., Chen, Y. & Yu, L.J. Uniformity of ionization response of structurally diverse analytes using a chip-based nanoelectrospray ionization source. Rapid Commun. Mass Spectrom. 19, 3139–3142 (2005).
Kind, T. & Fiehn, O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics 7, 234 (2006).
Sumner, L.W. et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 3, 211–221 (2007).
Davidson, R.L., Weber, R.J.M., Liu, H., Sharma-Oates, A. & Viant, M.R. Galaxy-M: a Galaxy workflow for processing and analysing direct infusion and liquid chromatography mass spectrometry-based metabolomics data. Gigascience 5, 10 (2015).
Kirwan, J.A., Weber, R.J.M., Broadhurst, D.I. & Viant, M.R. Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci. Data 1, 140012 (2014).
Hao, J. et al. Statistical correlations between NMR spectroscopy and direct infusion FT-ICR mass spectrometry aid annotation of unknowns in metabolomics. Anal. Chem. 88, 2583–2589 (2016).
Lin, L. et al. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 135, 2970–2978 (2010).
Annesley, T.M. Ion suppression in mass spectrometry. Clin. Chem. 49, 1041–1044 (2003).
Schmidt, A., Karas, M. & Dulcks, T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI? J. Am. Soc. Mass Spectrom. 14, 492–500 (2003).
Yang, K. & Han, X. Accurate quantification of lipid species by electrospray ionization mass spectrometry — meets a key challenge in lipidomics. Metabolites 1, 21–40 (2011).
Poynton, H.C. et al. Metabolomics of microliter hemolymph samples enables an improved understanding of the combined metabolic and transcriptional responses of Daphnia magna to cadmium. Environ. Sci. Tech. 45, 3710–3717 (2011).
Gonzalez-Dominguez, R., Castilla-Quintero, R., Garcia-Barrera, T. & Luis Gomez-Ariza, J. Development of a metabolomic approach based on urine samples and direct infusion mass spectrometry. Anal. Biochem. 465, 20–27 (2014).
Lokhov, P.G. et al. Prediction of classical clinical chemistry parameters using a direct infusion mass spectrometry. International Journal of Mass Spectrometry http://dx.doi.org/10.1016/j.ijms.2015.08.006 (2015).
Drury, N.E. et al. The effect of perhexiline on myocardial protection during coronary artery surgery: a two-centre, randomized, double-blind, placebo-controlled trial. Eur. J. Cardio-Thorac. Surg. 47, 464–472 (2015).
Mirbahai, L. et al. Disruption of DNA methylation via S-adenosylhomocysteine is a key process in high incidence liver carcinogenesis in fish. J. Proteome Res. 12, 2895–2904 (2013).
Weber, R.J.M., Selander, E., Sommer, U. & Viant, M.R. A stable-isotope mass spectrometry-based metabolic footprinting approach to analyze exudates from phytoplankton. Mar. Drugs 11, 4158–4175 (2013).
Southam, A.D. et al. Distinguishing between the metabolome and xenobiotic exposome in environmental field samples analysed by direct-infusion mass spectrometry based metabolomics and lipidomics. Metabolomics 10, 1050–1058 (2014).
Sturla, S.J. et al. Systems toxicology: from basic research to risk assessment. Chem. Res. Toxicol. 27, 314–329 (2014).
Van Aggelen, G. et al. Integrating omic technologies into aquatic ecological risk assessment and environmental monitoring: hurdles, achievements, and future outlook. Environ. Health Perspect. 118, 1–5 (2010).
Parsons, H.M., Ekman, D.R., Collette, T.W. & Viant, M.R. Spectral relative standard deviation: a practical benchmark in metabolomics. Analyst 134, 478–485 (2009).
Sellick, C.A., Hansen, R., Stephens, G.M., Goodacre, R. & Dickson, A.J. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat. Protoc. 6, 1241–1249 (2011).
Taylor, N.S., Weber, R.J.M., White, T.A. & Viant, M.R. Discriminating between different acute chemical toxicities via changes in the Daphnid metabolome. Toxicol. Sci. 118, 307–317 (2010).
Want, E.J. et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 8, 17–32 (2013).
Want, E.J. et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 5, 1005–1018 (2010).
Dunn, W.B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011).
Wu, H., Southam, A.D., Hines, A. & Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 372, 204–212 (2008).
Lin, C.Y., Wu, H.F., Tjeerdema, R.S. & Viant, M.R. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics 3, 55–67 (2007).
Han, X., Yang, K. & Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrome. Rev. 31, 134–178 (2012).
Bligh, E.G. & Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Matyash, V., Liebisch, G., Kurzchalia, T.V., Shevchenko, A. & Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008).
Schultz, G.A., Corso, T.N., Prosser, S.J. & Zhang, S. A fully integrated monolithic microchip electrospray device for mass spectrometry. Anal. Chem. 72, 4058–4063 (2000).
Zhang, L.K., Rempel, D., Pramanik, B.N. & Gross, M.L. Accurate mass measurements by Fourier transform mass spectrometry. Mass Spectrom. Rev. 24, 286–309 (2005).
Payne, T.G., Southam, A.D., Arvanitis, T.N. & Viant, M.R. A signal filtering method for improved quantification and noise discrimination in Fourier transform ion cyclotron resonance mass spectrometry-based metabolomics data. J. Am. Soc. Mass Spectrom. 20, 1087–1095 (2009).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006).
Kirwan, J.A., Broadhurst, D.I., Davidson, R.L. & Viant, M.R. Characterising and correcting batch variation in an automated direct infusion mass spectrometry (DIMS) metabolomics workflow. Anal. Bioanal. Chem. 405, 5147–5157 (2013).
Hrydziuszko, O. & Viant, M.R. Missing values in mass spectrometry based metabolomics: an undervalued step in the data processing pipeline. Metabolomics 8, S161–S174 (2012).
Parsons, H.M., Ludwig, C., Gunther, U.L. & Viant, M.R. Improved classification accuracy in 1-and 2-dimensional NMR metabolomics data using the variance stabilising generalised logarithm transformation. BMC Bioinformatics 8, 16 (2007).
van den Berg, R.A., Hoefsloot, H.C.J., Westerhuis, J.A., Smilde, A.K. & van der Werf, M.J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7, 142 (2006).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Good, P.L. Permutation, Parametric, and Bootstrap Tests of Hypotheses (Springer, 2005).
Szymanska, E., Saccenti, E., Smilde, A.K. & Westerhuis, J.A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 8, 3–16 (2012).
Dunn, W.B. et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 9, S44–S66 (2013).
Weber, R.J.M. & Viant, M.R. MI-Pack: increased confidence of metabolite identification in mass spectra by integrating accurate masses and metabolic pathways. Chemometrics Intell. Lab. Syst. 104, 75–82 (2010).
Kind, T. & Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 8, 105 (2007).
Haug, K. et al. MetaboLights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–D786 (2013).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2015).
Bennett, B.D., Yuan, J., Kimball, E.H. & Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 3, 1299–1311 (2008).
Taylor, N.S. et al. Metabolomics confirms that dissolved organic carbon mitigates copper toxicity. Environ. Toxicol. Chem. 35, 635–644 (2015).
Acknowledgements
A.D.S. was funded by a Bloodwise programme grant (no. 13028) awarded to F.L. Khanim and M.T. Drayson. R.J.M.W., M.R.V., J.E. and M.R.J. were funded by the UK Natural Environment Research Council (grants NE/I008314/1, NE/J017442/1, NE/K011294/1 and R8-H10-61), and R.J.M.W. and M.R.V. were funded by the UK Biotechnology and Biological Sciences Research Council (grant BB/M019985/1). M.R.J. thanks Thermo Fisher Scientific for CASE funding. The LTQ FT Ultra used in this research was obtained through the Birmingham Science City Translational Medicine: Experimental Medicine Network of Excellence project, with support from Advantage West Midlands (AWM). We thank N. Taylor (University of Birmingham, UK) and collaborators (J. Gunn, Laurentian University, Canada; and J. McGeer, Wilfrid Laurier University, Canada), who kindly provided us with experimental data for Figure 6. We thank D. Broadhurst (University of Alberta) for ongoing development and support for the batch-correction algorithm.
Author information
Authors and Affiliations
Contributions
A.D.S. and R.J.M.W. codeveloped the analytical methods and computational workflow, acquired data and wrote the manuscript. J.E. contributed to developing the computational workflow and writing of the manuscript. M.R.J. contributed to developing the analytical methods and acquired data. M.R.V. codeveloped the analytical methods and computational workflows, edited the manuscript and was the academic lead.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Methods 1
Q Exactive instrument method files. (ZIP 309 kb)
Supplementary Methods 2
Orbitrap Elite instrument method files. (ZIP 47 kb)
Rights and permissions
About this article
Cite this article
Southam, A., Weber, R., Engel, J. et al. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat Protoc 12, 310–328 (2017). https://doi.org/10.1038/nprot.2016.156
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.156
This article is cited by
-
Derivation of metabolic point of departure using high-throughput in vitro metabolomics: investigating the importance of sampling time points on benchmark concentration values in the HepaRG cell line
Archives of Toxicology (2023)
-
Spectral binning as an approach to post-acquisition processing of high resolution FIE-MS metabolome fingerprinting data
Metabolomics (2022)
-
Integrating in vitro metabolomics with a 96-well high-throughput screening platform
Metabolomics (2022)
-
An integrated host-microbiome response to atrazine exposure mediates toxicity in Drosophila
Communications Biology (2021)
-
Space and patchiness affects diversity–function relationships in fungal decay communities
The ISME Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.