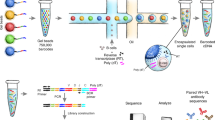
Abstract
High-throughput sequencing analysis of hypermutating immunoglobulin (IG) repertoires remains a challenging task. Here we present a robust protocol for the full-length profiling of human and mouse IG repertoires. This protocol uses unique molecular identifiers (UMIs) introduced in the course of cDNA synthesis to control bottlenecks and to eliminate PCR and sequencing errors. Using asymmetric 400+100-nt paired-end Illumina sequencing and UMI-based assembly with the new version of the MIGEC software, the protocol allows up to 750-nt lengths to be sequenced in an almost error-free manner. This sequencing approach should also be applicable to various tasks beyond immune repertoire studies. In IG profiling, the achieved length of high-quality sequence covers the variable region of even the longest chains, along with the fragment of a constant region carrying information on the antibody isotype. The whole protocol, including preparation of cells and libraries, sequencing and data analysis, takes 5 to 6 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Robins, H.S. et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114, 4099–4107 (2009).
Freeman, J.D., Warren, R.L., Webb, J.R., Nelson, B.H. & Holt, R.A. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 19, 1817–1824 (2009).
Mamedov, I.Z. et al. Quantitative tracking of T cell clones after haematopoietic stem cell transplantation. EMBO Mol. Med. 3, 201–207 (2011).
Warren, R.L. et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 21, 790–797 (2011).
Vollmers, C., Sit, R.V., Weinstein, J.A., Dekker, C.L. & Quake, S.R. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl. Acad. Sci. USA 110, 13463–13468 (2013).
Jiang, N. et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 5, 171ra119 (2013).
Laserson, U. et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc. Natl. Acad. Sci. USA 111, 4928–4933 (2014).
Kaplinsky, J. et al. Antibody repertoire deep sequencing reveals antigen-independent selection in maturing B cells. Proc. Natl. Acad. Sci. USA 111, E2622–2629 (2014).
Georgiou, G. et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158–168 (2014).
Weinstein, J.A., Jiang, N., White, R.A., Fischer, D.S. & Quake, S.R. High-throughput sequencing of the zebrafish antibody repertoire. Science 324, 807–810 (2009).
Mora, T., Walczak, A.M., Bialek, W. & Callan, C.G. Jr. Maximum entropy models for antibody diversity. Proc. Natl. Acad. Sci. USA 107, 5405–5410 (2010).
Jiang, N. et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. USA 108, 5348–5353 (2011).
Rubelt, F. et al. Onset of immune senescence defined by unbiased pyrosequencing of human immunoglobulin mRNA repertoires. PLoS One 7, e49774 (2012).
Parameswaran, P. et al. Convergent antibody signatures in human dengue. Cell Host Microbe 13, 691–700 (2013).
Tan, Y.C. et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin. Immunol. 151, 55–65 (2014).
Galson, J.D. et al. BCR repertoire sequencing: different patterns of B-cell activation after two Meningococcal vaccines. Immunol. Cell Biol. 93, 885–895 (2015).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K.W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 108, 9530–9535 (2011).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2012).
Britanova, O.V. et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 192, 2689–2698 (2014).
Shugay, M. et al. Towards error-free profiling of immune repertoires. Nat. Methods 11, 653–655 (2014).
He, L. et al. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci. Rep. 4, 6778 (2014).
Egorov, E.S. et al. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J. Immunol. 194, 6155–6163 (2015).
Khan, T.A. et al. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci. Adv. 2, e1501371 (2016).
Bolotin, D.A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
Briney, B.S., Willis, J.R., McKinney, B.A. & Crowe, J.E. Jr. High-throughput antibody sequencing reveals genetic evidence of global regulation of the naive and memory repertoires that extends across individuals. Genes Immun. 13, 469–473 (2012).
Larimore, K., McCormick, M.W., Robins, H.S. & Greenberg, P.D. Shaping of human germline IgH repertoires revealed by deep sequencing. J. Immunol. 189, 3221–3230 (2012).
Wu, Y.C. et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 116, 1070–1078 (2010).
Siegrist, C.A. & Aspinall, R. B-cell responses to vaccination at the extremes of age. Nat. Rev. Immunol. 9, 185–194 (2009).
Wang, C. et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 192, 603–611 (2014).
Tan, Y.C. et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 66, 2706–2715 (2014).
Doorenspleet, M.E. et al. Rheumatoid arthritis synovial tissue harbours dominant B-cell and plasma-cell clones associated with autoreactivity. Ann. Rheum. Dis. 73, 756–762 (2014).
Racanelli, V. et al. Antibody V(h) repertoire differences between resolving and chronically evolving hepatitis C virus infections. PLoS One 6, e25606 (2011).
Ademokun, A. et al. Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell 10, 922–930 (2011).
Tschumper, R.C. et al. Comprehensive assessment of potential multiple myeloma immunoglobulin heavy chain V-D-J intraclonal variation using massively parallel pyrosequencing. Oncotarget 3, 502–513 (2012).
Fridy, P.C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014).
Lu, D.R. et al. Identifying functional anti-Staphylococcus aureus antibodies by sequencing antibody repertoires of patient plasmablasts. Clin. Immunol. 152, 77–89 (2014).
Lavinder, J.J. et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. USA 111, 2259–2264 (2014).
Briney, B.S., Willis, J.R. & Crowe, J.E. Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One 7, e36750 (2012).
Yu, L. & Guan, Y. Immunologic basis for long HCDR3s in broadly neutralizing antibodies against HIV-1. Front. Immunol. 5, 250 (2014).
Nguyen, P. et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics 12, 106 (2011).
Bolotin, D.A. et al. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur. J. Immunol. 42, 3073–3083 (2012).
Brodin, J. et al. PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data. PLoS One 8, e70388 (2013).
Brodin, J. et al. Challenges with using primer IDs to improve accuracy of next generation sequencing. PLoS One 10, e0119123 (2015).
Yaari, G. & Kleinstein, S.H. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. 7, 121 (2015).
Elnifro, E.M., Ashshi, A.M., Cooper, R.J. & Klapper, P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13, 559–570 (2000).
Markoulatos, P., Siafakas, N. & Moncany, M. Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal. 16, 47–51 (2002).
Carlson, C.S. et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 4, 2680 (2013).
van Dijk, E.L., Jaszczyszyn, Y. & Thermes, C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322, 12–20 (2014).
Schrum, A.G., Turka, L.A. & Palmer, E. Surface T-cell antigen receptor expression and availability for long-term antigenic signaling. Immunol. Rev. 196, 7–24 (2003).
Cho, B.K., Wang, C., Sugawa, S., Eisen, H.N. & Chen, J. Functional differences between memory and naive CD8 T cells. Proc. Natl. Acad. Sci. USA 96, 2976–2981 (1999).
Schrum, A.G., Wells, A.D. & Turka, L.A. Enhanced surface TCR replenishment mediated by CD28 leads to greater TCR engagement during primary stimulation. Int. Immunol. 12, 833–842 (2000).
Shi, W. et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16, 663–673 (2015).
Wrammert, J. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008).
Franz, B., May, K.F. Jr., Dranoff, G. & Wucherpfennig, K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood 118, 348–357 (2011).
Kuenz, B. et al. Cerebrospinal fluid B cells correlate with early brain inflammation in multiple sclerosis. PLoS One 3, e2559 (2008).
Greiff, V. et al. Quantitative assessment of the robustness of next-generation sequencing of antibody variable gene repertoires from immunized mice. BMC Immunology 15, 40 (2014).
Matz, M. et al. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27, 1558–1560 (1999).
Douek, D.C. et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168, 3099–3104 (2002).
Feng, Y. et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature 528, 132–136 (2015).
Shugay, M. et al. VDJtools: unifying post-analysis of T cell receptor repertoires. PLoS Comput. Biol. 11, e1004503 (2015).
Jaatinen, T. & Laine, J. Isolation of mononuclear cells from human cord blood by Ficoll-Paque density gradient. Curr. Protoc. Stem Cell Biol. Chapter 2 Unit 2A 1, http://dx.doi.org/10.1002/9780470151808.sc02a01s1 (2007).
Sims, G.P. & Lipsky, P.E. Isolation of human B cell populations. Curr. Protoc. Immunol. Chapter 7 Unit 7 5, http://dx.doi.org/10.1002/0471142735.im0705s75 (2006).
Kjeldsen, M.K. et al. Multiparametric flow cytometry for identification and fluorescence activated cell sorting of five distinct B-cell subpopulations in normal tonsil tissue. Am. J. Clin. Pathol. 136, 960–969 (2011).
Acknowledgements
This work was supported by Russian Science Foundation project no. 14-14-00533. The work was carried out in part using equipment provided by the Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry Core Facility (CKP IBCH). M.S. is supported by an individual fellowship (mol-a-dk RFBR grant 16-34-60179). A.D., V.B., K.P. and S.P. are supported by the Ministry of Education, Youth and Sports of the Czech Republic (CEITEC 2020, LQ1601). K.P., V.B. and S.P. are supported by Ministry of Health, Czech Republic (AZV 15-30015A).
Author information
Authors and Affiliations
Contributions
M.A.T., O.V.B., V.B., V.I.K., E.M.M., I.Z.M. and K.P. prepared the cDNA libraries and worked on the protocol. E.S.E., D.B.S. and O.K. worked on cell sample preparation. A.D. and M.D.L. worked on sequencing. M.A.T., O.V.B. and D.M.C. designed the experiments. A.D., M.S., D.A.B., M.I., S.P. and D.M.C. worked on data analysis and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Title: Experimental datasets. Examples of full-length IGH profiling for the samples of human naive, plasma and memory B cells (XLSX 11235 kb)
Supplementary Table 2
Title: Control datasets. Full-length IGH profiling for the control set of CLL clones (XLSX 50 kb)
Rights and permissions
About this article
Cite this article
Turchaninova, M., Davydov, A., Britanova, O. et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc 11, 1599–1616 (2016). https://doi.org/10.1038/nprot.2016.093
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.093
This article is cited by
-
Current advancements in B-cell receptor sequencing fast-track the development of synthetic antibodies
Molecular Biology Reports (2024)
-
A dual function for the chromatin organizer Special A-T rich Binding Protein 1 in B-lineage cells
Cellular & Molecular Immunology (2023)
-
Molecular characterization stratifies VQ myeloma cells into two clusters with distinct risk signatures and drug responses
Oncogene (2023)
-
Origin and evolutionary malleability of T cell receptor α diversity
Nature (2023)
-
Longitudinal analyses of CLL in mice identify leukemia-related clonal changes including a Myc gain predicting poor outcome in patients
Leukemia (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.